Entering edit mode
Hello,
I am working on two Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray datasets. One dataset only includes metastatic samples to the A site, and the other includes metastatic samples to the B site only. I merged this dataset to find differentially expressed genes. I plot PCA, to check batch effect, and I added PCA graph below. My two biological group coming from two different dataset. How can ı remove batch effect?

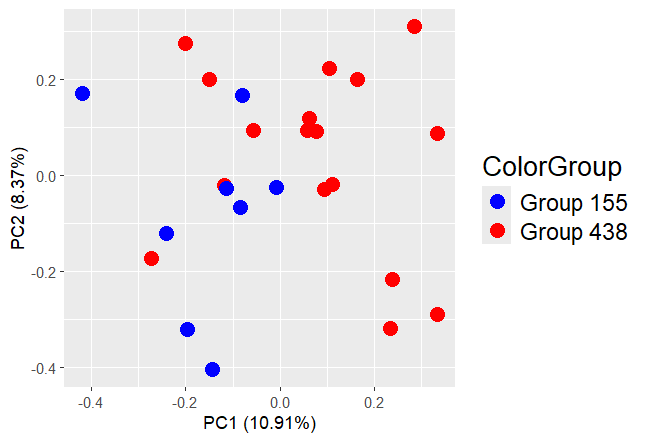

Thank you
Hello again, Can using inSilicoMerging handle this situation?