Entering edit mode
Hi, I am doing RNAseq analysis on various tissues such as placenta & cord blood, parent blood. There are case control groups (each around 40) based on health outcomes of infants and no biological duplicate of samples.
- After quantifying through salmon, I want to know what is the best approach for proceeding for Differentially expressed genes? I am doing separate analysis for each tissue type till the significant list of DEGs of a group and then comparison or correlation of these genes with each of the tissue type i.e., DEGs of case control from placenta with that of cord blood. Also I will compare them at functional levels.
- How can I compare gene expression pattern of parents with infants?
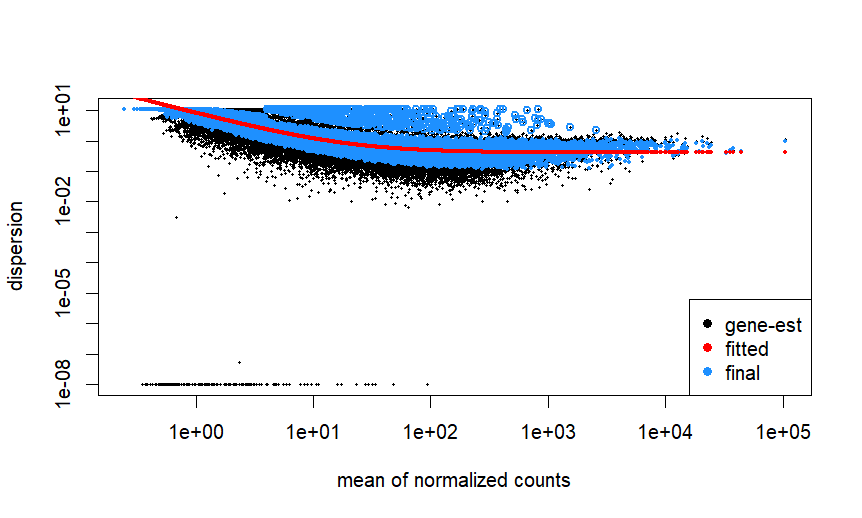
- can I process via DESeq2 as the dispersion for 10 pilot samples looks fine if I am not wrong.
Thank you so much.