Dear community.
on my way to address alternatively used exons in a bulkRNAseq dataset I used fastq files and aligned reads with Rsubjunc (see in code below).
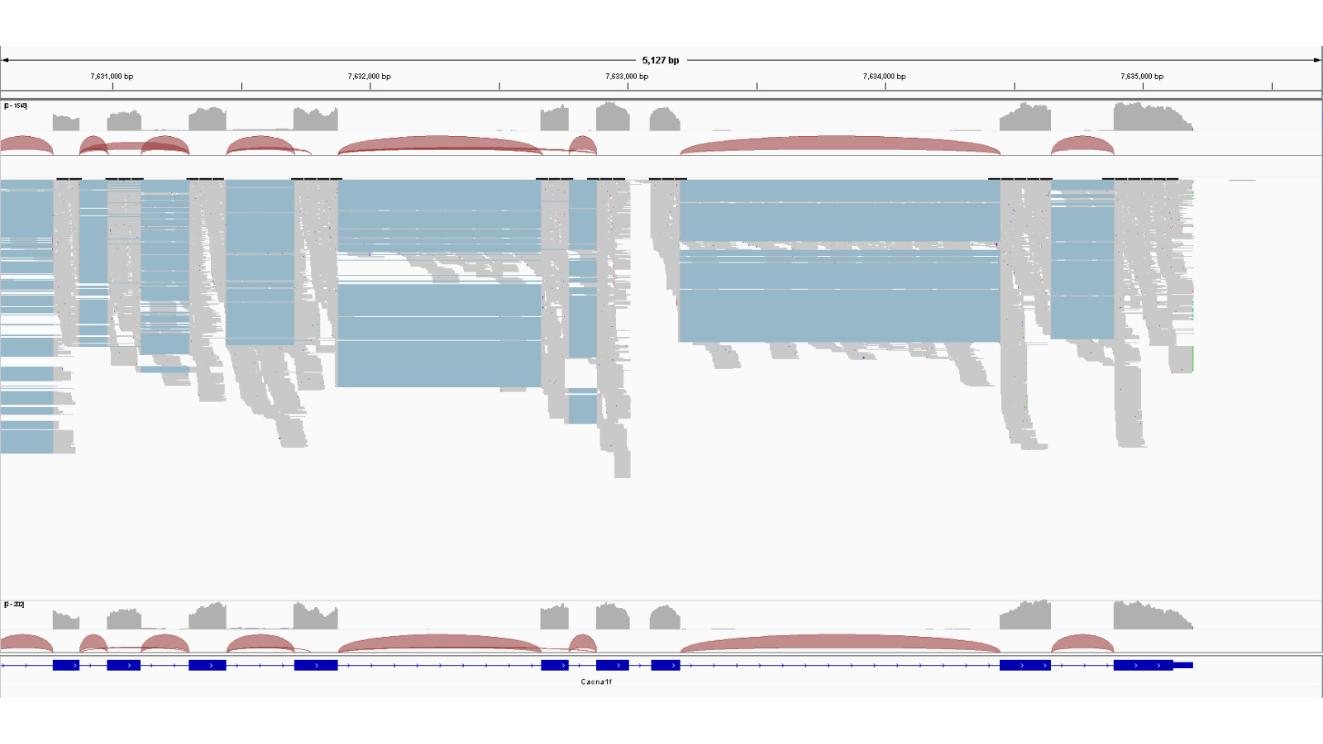
To get a first impression I fed in the bam files of our control into IGV and looked a gene I know from from earlier studies and was surprised to see that the end of the transcript is split, as if the C-terminus was not connected on mRNA level. This confused be because the mRNA should be connected. Is there something, a setting, or something else I am missing here?
I merged all six bam files in hope the merged file get more read connecting Ex45 with Ex46 (Total exon count is 48).
Additionally, quiet a few reads are also found in the intronic regions, although not connected enough to say this is intron retention?
I am happy to hear your opinion on this.
fastq_files <- list.files("../fastq/raw/", full.names = T, include.dirs = F)
fastq_files_abs <- normalizePath(fastq_files)
subjunc(index = "/home/chuddy/bioinformatics/ref_genomes/mouse_38/genome/subread/GRCm38",
nthreads = 20,
readfile1 = fastq_files_abs,
sortReadsByCoordinates = TRUE)
> sessionInfo( )
R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=de_CH.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=de_CH.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=de_CH.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=de_CH.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Zurich
tzcode source: system (glibc)
attached base packages:
[1] grid stats graphics grDevices utils datasets methods base
other attached packages:
[1] gridExtra_2.3 Rsubread_2.14.2 lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[8] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] Matrix_1.6-5 gtable_0.3.4 compiler_4.3.2 tidyselect_1.2.0 scales_1.3.0 lattice_0.22-5
[7] R6_2.5.1 generics_0.1.3 knitr_1.45 munsell_0.5.0 pillar_1.9.0 tzdb_0.4.0
[13] rlang_1.1.3 utf8_1.2.4 stringi_1.8.3 xfun_0.41 timechange_0.3.0 cli_3.6.2
[19] withr_3.0.0 magrittr_2.0.3 rstudioapi_0.15.0 hms_1.1.3 lifecycle_1.0.4 vctrs_0.6.5
[25] glue_1.7.0 fansi_1.0.6 colorspace_2.1-0 tools_4.3.2 pkgconfig_2.0.3