Entering edit mode
I have been trying to install rhdf5 in Rstudio which is using the R version 4.1.3 so that can use MplusAutomation. I have successfully installed BiocManager 1.30.21 but it does not allow me to install rhdf5 as it indicates that this package is not available for Bioconductor version "3.14".
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("rhdf5")
sessionInfo( )
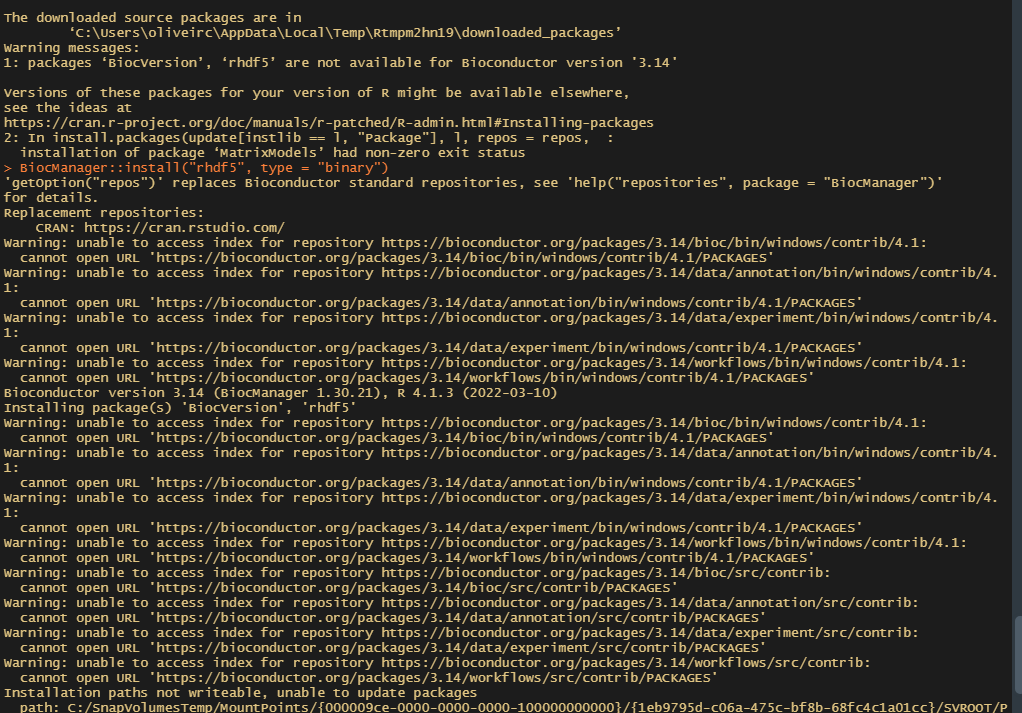
Sorry I had to post screenshots, I am working on a controlled server that does not allow me to copy.
Thank you!


This doesn't look like an rhdf5 issue, but more a general internet connectivity problem. You would not expect to see all those "cannot open URL" messages if everything was working smoothly.
If you're working on a locked down environment where you can't copy, is it also possible that there's a firewall preventing you connecting to the Bioconductor servers? It might also be worth noting that Bioc 3.14 is fairly old and has been moved to some archive storage. It's still available, but the URLs will redirect somewhere different to get it e.g.
https://bioconductor.org/packages/3.14/bioc/bin/windows/contrib/4.1/PACKAGESredirects to
https://mghp.osn.xsede.org/bir190004-bucket01/archive.bioconductor.org/packages/3.14/bioc/bin/windows/contrib/4.1/PACKAGESIf you need to open a firewall, you'll probably want to make sure the second address is also opened.