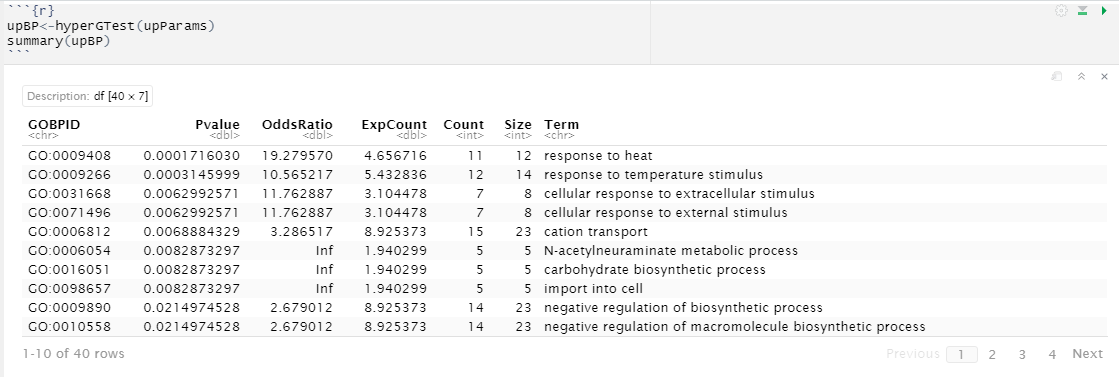
Hi I am very new to this project and I am not sure How to interpret the gene enrichment analysis result. What does each column mean? See the picture in comment ```
Gene ontology enrichment analysis
0
Entering edit mode
0
Entering edit mode
ADD REPLY
• link
2.4 years ago
acram
•
0
0
Entering edit mode
The columns are, in order
- The GO ID
- The p-value for the hypergeometric test of over-representation
- The odds ratio. This isn't particularly useful IMO, but it's the odds of being significant, given that a gene is in the GO term, divided by the odds of not being significant, given the gene is in the term. A large value indicates that it's way more likely for a gene that is in a given GO term to be significant than not.
- The expected number of significant genes for the GO term, under the null distribution (e.g., for the first row in your table, we expect around five significant genes for that GO term if we randomly select N genes to be significant, where N is the number of significant genes you observed).
- The observed count of significant genes for the given GO term
- The total number of genes for that GO term
- The descriptive name for the GO term
0
Entering edit mode
Further to the interpretation of the odds ratio. This comes from case/control studies, where you can set up a 2x2 table. So for example, you can do a study where you compare smokers to non-smokers and how many get lung cancer. The odds ratio in that context would compare the odds of getting lung cancer, given you are a smoker, to the odds of getting lung cancer for non-smokers. A large OR may indicate a relationship between lung cancer and smoking. In the context of a GO hypergeometric it's a less useful statistic, and mostly people just rely on the p-value.
Similar Posts
Loading Similar Posts
Traffic: 1076 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.