Dear All,
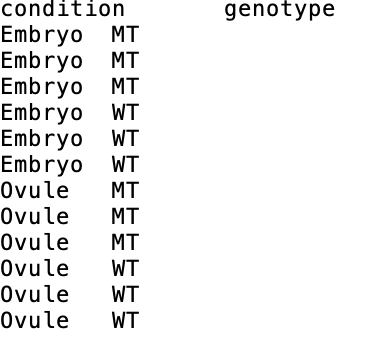
I have the samples of two cell types (Embryo and Ovule) with two genotypes (WT and MT), which indicate as condition and genotype below.
I would like to compare:
- EmbryoWT vs EmbryoMT,
- OvuleWT vs OvuleMT.
- EmbryoWT vs OvuleWT
- EmbryoMT vs OvuleMT.
I used this model to compare my samples and try to look for solutions. However, It seems to be the analysis will include the same condition or genotype together which is not our interest.
dds <- DESeqDataSetFromMatrix(countData = clean_data, colData = meta, design = ~ genotype + condition + genotype:condition)
I highly appreciate your help.
Thanks in advance,
Enter the body of text here
Code should be placed in three backticks as shown below
# include your problematic code here with any corresponding output
# please also include the results of running the following in an R session
sessionInfo( )


Has been cross-posted: https://www.biostars.org/p/9467526/