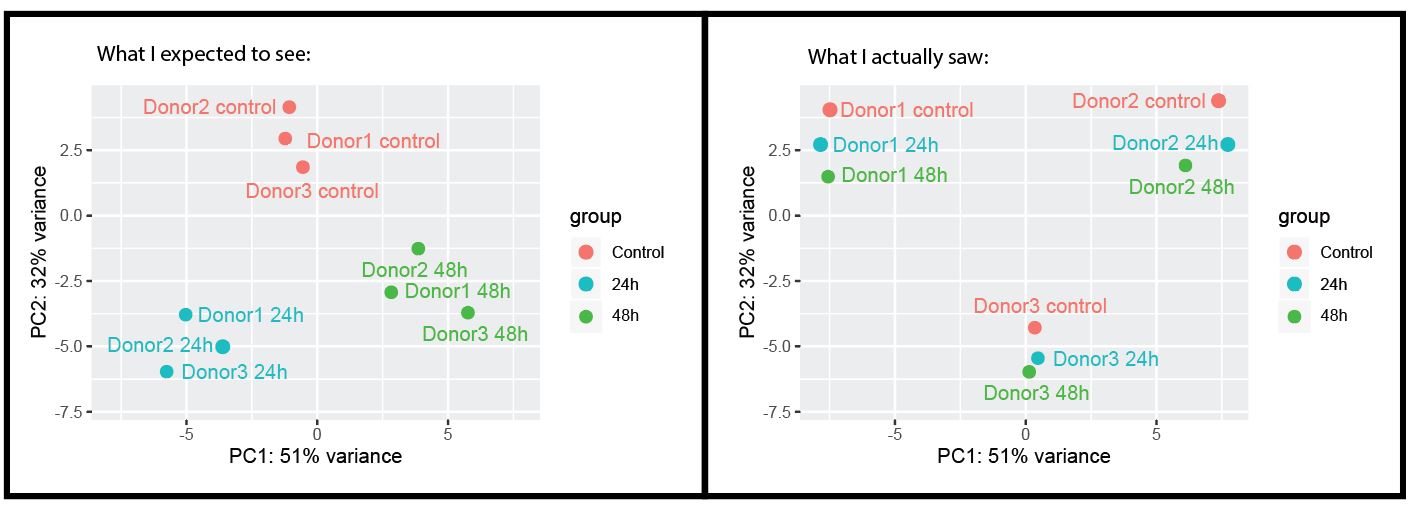
Hi! I want to perform time course analysis on my dataset. I have harvested samples from 3 intestinal organoid lines (which are derived from different patients) over 3 different timepoints (0h, 24h and 48h) after exposure to the compound. Now I have a bulk RNA-seq dataset, where I initially thought I could use the 3 donors as biological replicates, but it seems that the inter-patient differences are bigger than the effect that the treatment induces.
Just to illustrate:
So, since I don't have 3 technical replicates per donor for each timepoint (but each timepoint has 3 donors that I though I could use as biological replicates), I feel a bit stuck, because averaging over all biological donors will not give good results (because baselevels are different).
What I think I should do, is first fit a linear regression line on expression vs time for each donor separately and then compare those slopes and regard them as replicates? I am quite new to DESeq2 model design that involves interactions, so I was wondering if you could help me out?
Here's an overview about the components of my design:
Time = as.factor(c(0, 0, 0, 24, 24, 24, 48, 48, 48))
Donor = as.factor(c(1, 2, 3, 1, 2, 3, 1, 2, 3))
metadata = c("Time", "Donor")
After googling around a bit, I tried something like this:
dds <- DESeqDataSetFromMatrix(countData = myData, colData = metadata, design = ~Donor + Time + Donor:Time)
dds <- DESeq(dds, test = "LRT", reduced = ~ Donor + Time)
It started off fine, but then returned me an error:
Error in checkForExperimentalReplicates(object, modelMatrix) :
The design matrix has the same number of samples and coefficients to fit, so estimation of dispersion is not possible. Treating samples as replicates was deprecated in v1.20 and no longer supported since v1.22.
Do you have any suggestions what to change in design? And how to make sure that timepoint 0 is considered as a reference level? Thanks in advance!