I'm running DESeq2 on RNAseq data and my experimental design has three factors: Carbon_Source, Time_Point, and Amoxicillin. I'm having trouble extracting the comparisons that I need.
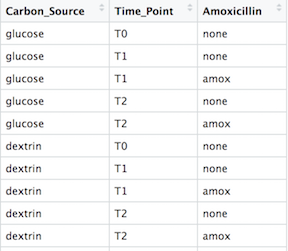
Here is a portion of my sample metadata:
dds = DESeqDataSetFromMatrix(control_table, Design, ~Carbon_Source + Time_Point + Amoxicillin + Carbon_Source:Amoxicillin)
dds$Carbon_Source = factor(dds$Carbon_Source, levels=c("glucose", "dextrin"))
dds$Time_Point = factor(dds$Time_Point, levels=c("T0", "T1", "T2"))
dds$Amoxicillin = factor(dds$Amoxicillin, levels=c("none", "amox"))
DDS <- DESeq(dds)
resultsNames(DDS)
[1] "Intercept" "Carbon_Source_dextrin_vs_glucose"
[3] "Time_Point_T1_vs_T0" "Time_Point_T2_vs_T0"
[5] "Amoxicillin_amox_vs_none" "Carbon_Sourcedextrin.Amoxicillinamox"
These are the comparisons that I think I have correct:
#Is the effect of amoxicillin different depending on glucose vs dextrin? (across timepoints)
res1 = results(DDS, name = "Carbon_Sourcedextrin.Amoxicillinamox")
#The effect of amoxicillin on the glucose condition:
res2 = results(DDS, name = "Amoxicillin_amox_vs_none")
#The effect of amoxicillin on the dextrin condition:
res3 = results(DDS, list( c("Amoxicillin_amox_vs_none", "Carbon_Sourcedextrin.Amoxicillinamox") ))
#Difference between glucose and dextrin conditions without amoxicillin:
res4 = results(DDS, name = "Carbon_Source_dextrin_vs_glucose")
#Difference between glucose T1 without amoxicillin and glucose T0:
res5 = results(DDS, name = "Time_Point_T1_vs_T0")
#Difference between glucose T2 without amoxicillin and glucose T0:
res6 = results(DDS, name = "Time_Point_T2_vs_T0")
I would also like to have the following comparisons, but I'm not sure how this can be done:
- Difference between glucose T1 with amoxicillin and glucose T0
- Difference between glucose T2 with amoxicillin and glucose T0
- Difference between dextrin T1 with amoxicillin and dextrin T0
- Difference between dextrin T2 with amoxicillin and dextrin T0
- Difference between glucose and dextrin conditions with amoxicillin
I'm not sure that these comparisons are even possible since T0 only had one treatment type (no amoxicillin). This is also why I can't do an interaction term with time_point because the model matrix would not be full rank.
One idea I had was to run DESeq2 again with the same design, except flip the baseline levels so that "dextrin" is the baseline for Carbon_Source and "amox" is the baseline for Amoxicillin.
Any advice is appreciated!

