Hello,
I have RNA-seq data from testis tissue of a natural population of 3 closely related bird species (5 individuals each).
I have detected several thousands (3195) differentially expressed genes in one of my comparisons between the two groups. When looking at some of the differentially expressed genes, I see quite high variation within one of my groups as shown for one example gene below. I have 5 individuals per each group. As can be seen, the group to the right, shows a high level of variation for this gene.

Not all genes show the above pattern. There are many like the one below as well.
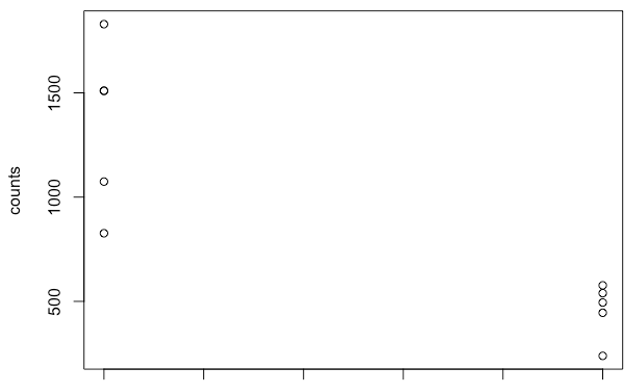
My question is that whether in the cases like the first image with large within-group variation, we can really say that the two groups differ and the gene is being differentially expressed since it looks as if there is one individual which has a very high expression value?
Thank you!


I understand better now. This issue has actually been directly addressed in Deseq2 paper in the section in which the authors talk about shrinkage of log2fold changes in lowly expressed genes (Love et al 2014). When I shrink the log2 fold change for the first case, the log2fold change goes from 6 to 0.23 which would filter out during my analysis. In the second cases, however, the log2fold change does not really change.