Hi,
I hope to try a full library size normalization but with edgeR.
So I performed the following:
data(tamoxifen_analysis)
tamoxifen = dba.normalize(tamoxifen, method = DBA_EDGER)
dba.normalize(tamoxifen, method = DBA_EDGER, bRetrieve = T)
Interestingly the normalization factors are all 1
$norm.method
[1] "lib"
$norm.factors
[1] 1 1 1 1 1 1 1 1 1 1 1
$lib.method
[1] "full"
$lib.sizes
[1] 652697 663370 346429 368052 466273 399879 1475415 616630 593224 706836 2575408
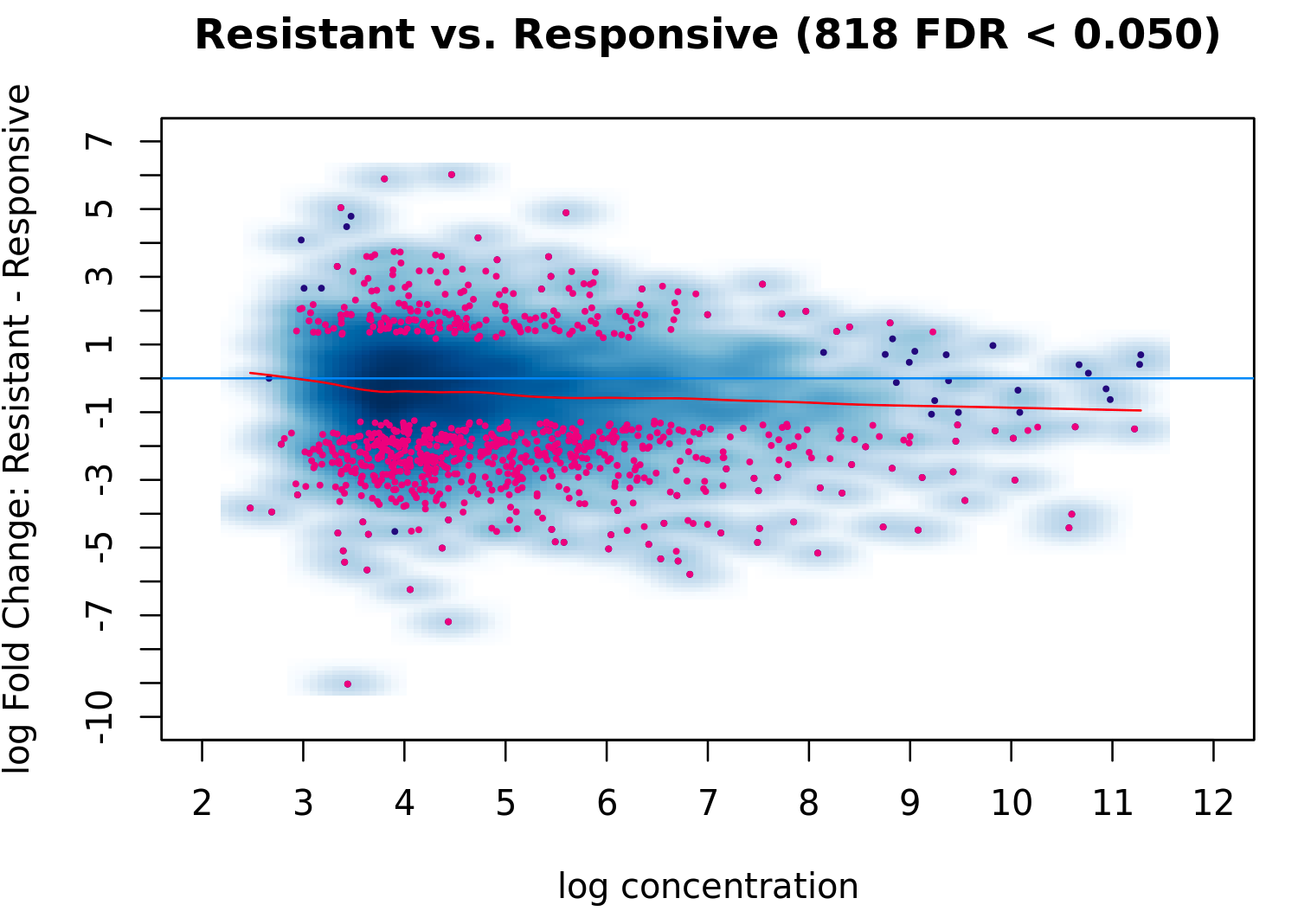
But apparently some normalization has happened as the ma plot is different from the one using raw data:
tamoxifen = dba.analyze(tamoxifen, method = DBA_EDGER)
dba.plotMA(tamoxifen, method = DBA_EDGER)
Could you please explain why is that? Thanks!


Hi, thanks for your response.
I'd like to know why
Shouldn't they be proportional to the actual library sizes?
I didn't want to do the more elaborate edgeR normalization, I just want to try the option of full library size + egdeR. In the tutorial, the author showed full library size + Deseq2 as the first and simplest normalization, where the norm factors are as expected (proportional to library sizes).
No, normalization factors from edgeR rather than size factors from DESeq2 are not proportional, it's a different concept and implementation of the same end goal. The edgeR normalization factors are as expected.