Hi all,
I have an experiment where I am interested in exploring the effects of three main factors: cell type, condition and disease (group), therefore I have a model that is ~ [covariates] + Type * Condition * Group.
I am following the suggestions I saw on this post, since it's very similar to what I have: Complex Contrast in DESeq2
However, I am still quite confused on how to select the right coefficients for which I want the results. Below is a description of each factor in my model:
- Type: Organoids, Organoids.CC, IELs, IEL.CC
- Condition: BDO.EM RBDO.IL2.7, RBDO.IL2.7.IFNy, RBDO.IL2.7.IL15.21. SAB.IL2
- Group: Ctrl, CeD
And these are the coefficients that I get from resultsNames(dds):
[1] "Intercept" "Age" "Sex_M_vs_F"
[4] "GC" "Rin" "QC_score_result"
[7] "Type_IELs.CC_vs_IELs" "Type_Organoids_vs_IELs" "Type_Organoids.CC_vs_IELs"
[10] "Condition_RBDO.IL2.7_vs_BDO.EM" "Condition_RBDO.IL2.7.IFNy_vs_BDO.EM" "Condition_RBDO.IL2.7.IL15.21_vs_BDO.EM"
[13] "Condition_SAB.IL2_vs_BDO.EM" "Group_CeD_vs_Ctrl" "TypeIELs.CC.ConditionRBDO.IL2.7"
[16] "TypeOrganoids.ConditionRBDO.IL2.7" "TypeOrganoids.CC.ConditionRBDO.IL2.7" "TypeIELs.CC.ConditionRBDO.IL2.7.IFNy"
[19] "TypeOrganoids.ConditionRBDO.IL2.7.IFNy" "TypeOrganoids.CC.ConditionRBDO.IL2.7.IFNy" "TypeIELs.CC.ConditionRBDO.IL2.7.IL15.21"
[22] "TypeOrganoids.ConditionRBDO.IL2.7.IL15.21" "TypeOrganoids.CC.ConditionRBDO.IL2.7.IL15.21" "TypeIELs.CC.ConditionSAB.IL2"
[25] "TypeOrganoids.ConditionSAB.IL2" "TypeOrganoids.CC.ConditionSAB.IL2" "TypeIELs.CC.GroupCeD"
[28] "TypeOrganoids.GroupCeD" "TypeOrganoids.CC.GroupCeD" "ConditionRBDO.IL2.7.GroupCeD"
[31] "ConditionRBDO.IL2.7.IFNy.GroupCeD" "ConditionRBDO.IL2.7.IL15.21.GroupCeD" "ConditionSAB.IL2.GroupCeD"
[34] "TypeIELs.CC.ConditionRBDO.IL2.7.GroupCeD" "TypeOrganoids.ConditionRBDO.IL2.7.GroupCeD" "TypeOrganoids.CC.ConditionRBDO.IL2.7.GroupCeD"
[37] "TypeIELs.CC.ConditionRBDO.IL2.7.IFNy.GroupCeD" "TypeOrganoids.ConditionRBDO.IL2.7.IFNy.GroupCeD" "TypeOrganoids.CC.ConditionRBDO.IL2.7.IFNy.GroupCeD"
[40] "TypeIELs.CC.ConditionRBDO.IL2.7.IL15.21.GroupCeD" "TypeOrganoids.ConditionRBDO.IL2.7.IL15.21.GroupCeD" "TypeOrganoids.CC.ConditionRBDO.IL2.7.IL15.21.GroupCeD"
[43] "TypeIELs.CC.ConditionSAB.IL2.GroupCeD" "TypeOrganoids.ConditionSAB.IL2.GroupCeD" "TypeOrganoids.CC.ConditionSAB.IL2.GroupCeD"
I found a bit challenging to understand how I can select the correct coefficients. For example, let's say I want to compare Organoids.CC in RBDO.IL2.7.IL15.21 GroupCeD with Organoids.CC in the same media but Group Control. How would I specify that in the coef parameter of results() or lfcShrink()?
Another approach I tried was building a contrast using the following code. Let's assume I am interested in the comparison "Type_Organoids_vs_IELs" (global difference between Organoids and IELs, regardless of condition and group), since this is a easy one to get using the coeficients way. Then, I would build the contrast like this:
# Build the contrast matrix
mod_mat <- model.matrix(design(dds), colData(dds))
# Select the right contrast
Type1 <- 'IELs'
Type2 <- 'Organoids'
C1 <- colMeans(mod_mat[dds$Type == Type1,])
C2 <- colMeans(mod_mat[dds$Type == Type2,])
contrast <- C2 - C1
Where print(contrast) is:
(Intercept) Age SexM
0.000000e+00 -7.438494e-17 0.000000e+00
GC Rin QC_score_result
6.823536e-02 2.371657e-01 -5.149269e-01
TypeIELs.CC TypeOrganoids TypeOrganoids.CC
0.000000e+00 1.000000e+00 0.000000e+00
ConditionRBDO.IL2.7 ConditionRBDO.IL2.7.IFNy ConditionRBDO.IL2.7.IL15.21
0.000000e+00 0.000000e+00 0.000000e+00
ConditionSAB.IL2 GroupCeD TypeIELs.CC:ConditionRBDO.IL2.7
0.000000e+00 0.000000e+00 0.000000e+00
TypeOrganoids:ConditionRBDO.IL2.7 TypeOrganoids.CC:ConditionRBDO.IL2.7 TypeIELs.CC:ConditionRBDO.IL2.7.IFNy
2.000000e-01 0.000000e+00 0.000000e+00
TypeOrganoids:ConditionRBDO.IL2.7.IFNy TypeOrganoids.CC:ConditionRBDO.IL2.7.IFNy TypeIELs.CC:ConditionRBDO.IL2.7.IL15.21
2.000000e-01 0.000000e+00 0.000000e+00
TypeOrganoids:ConditionRBDO.IL2.7.IL15.21 TypeOrganoids.CC:ConditionRBDO.IL2.7.IL15.21 TypeIELs.CC:ConditionSAB.IL2
2.000000e-01 0.000000e+00 0.000000e+00
TypeOrganoids:ConditionSAB.IL2 TypeOrganoids.CC:ConditionSAB.IL2 TypeIELs.CC:GroupCeD
2.000000e-01 0.000000e+00 0.000000e+00
TypeOrganoids:GroupCeD TypeOrganoids.CC:GroupCeD ConditionRBDO.IL2.7:GroupCeD
5.000000e-01 0.000000e+00 0.000000e+00
ConditionRBDO.IL2.7.IFNy:GroupCeD ConditionRBDO.IL2.7.IL15.21:GroupCeD ConditionSAB.IL2:GroupCeD
0.000000e+00 0.000000e+00 0.000000e+00
TypeIELs.CC:ConditionRBDO.IL2.7:GroupCeD TypeOrganoids:ConditionRBDO.IL2.7:GroupCeD TypeOrganoids.CC:ConditionRBDO.IL2.7:GroupCeD
0.000000e+00 1.000000e-01 0.000000e+00
TypeIELs.CC:ConditionRBDO.IL2.7.IFNy:GroupCeD TypeOrganoids:ConditionRBDO.IL2.7.IFNy:GroupCeD TypeOrganoids.CC:ConditionRBDO.IL2.7.IFNy:GroupCeD
0.000000e+00 1.000000e-01 0.000000e+00
TypeIELs.CC:ConditionRBDO.IL2.7.IL15.21:GroupCeD TypeOrganoids:ConditionRBDO.IL2.7.IL15.21:GroupCeD TypeOrganoids.CC:ConditionRBDO.IL2.7.IL15.21:GroupCeD
0.000000e+00 1.000000e-01 0.000000e+00
TypeIELs.CC:ConditionSAB.IL2:GroupCeD TypeOrganoids:ConditionSAB.IL2:GroupCeD TypeOrganoids.CC:ConditionSAB.IL2:GroupCeD
0.000000e+00 1.000000e-01 0.000000e+00
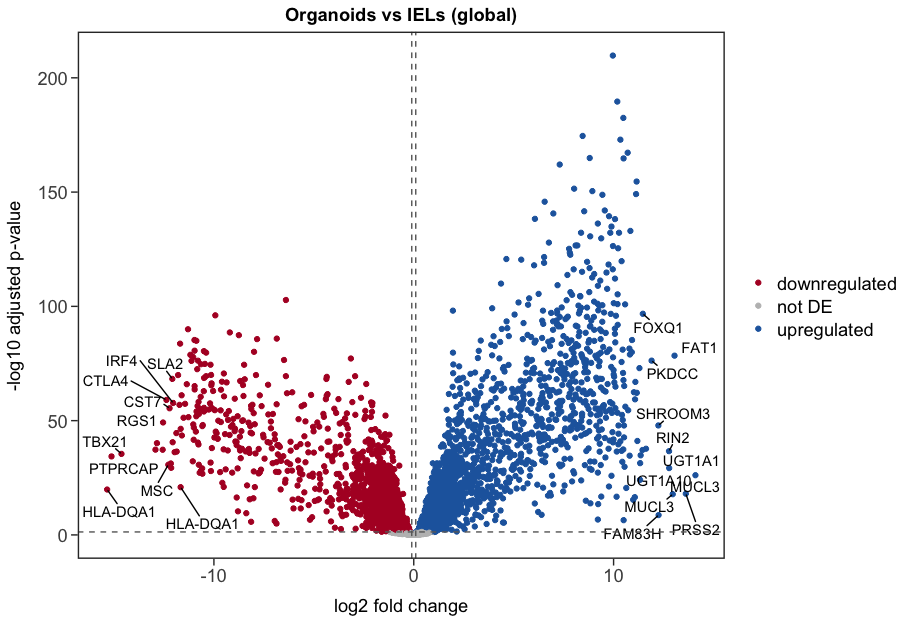
However, passing this contrast to lfcShrink, does not give me the same results as if I use coef Type_Organoids_vs_IELs, as shown below (1st plot is using contrast, 2nd is using coef):
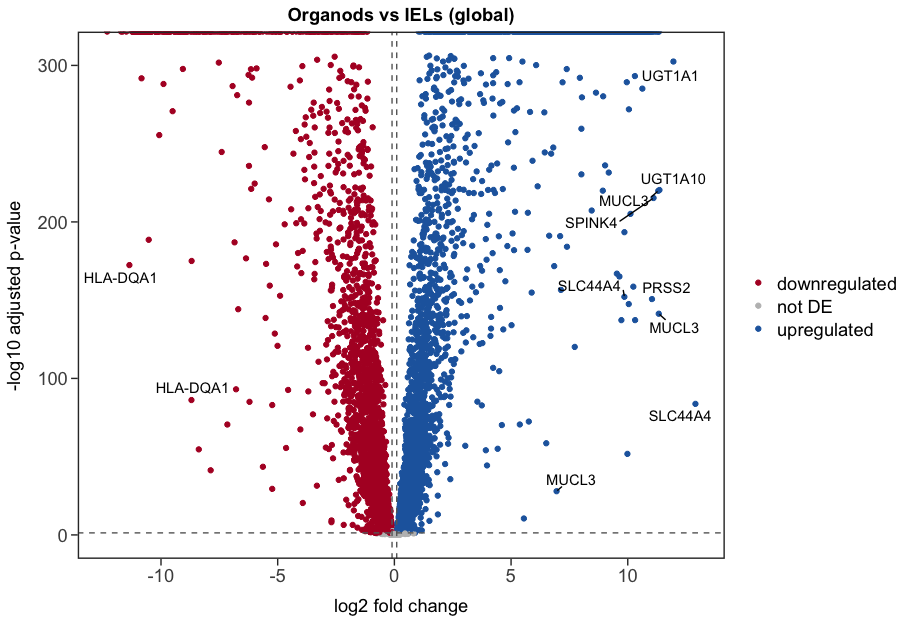
The results using contrast look very weird, with many pval=0, which goes to Inf in the log scale. That suggests me I am doing something wrong when using contrast.
I would really appreciate if someone could guide me a bit more on how to select the appropriate coefs in these situations where I want specific groups, types and conditions. Or if I can't do this using the coefs, how to properly do it with contrasts.
Thank you very much!