In a RNA-seq dataset, I have two batches that have one sample in common, plus other samples in each batch. There are 3 replicates for each sample. Is there a way to compare other samples across tthe two batches? In other words, how can the batch effects be removed?
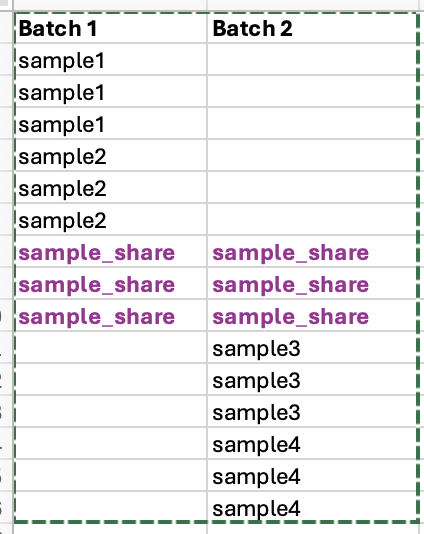
Is there a way to do DEG analysis across two batches sharing one sample only?
0
Entering edit mode
0
Entering edit mode
The short answer is yes, you can use even a single sample to estimate the batch effect (and therefore control for it in otherwise confounded comparison).
However you have low power with a single sample, as compared to an orthogonal (balanced) design. This is because there is substantial variance from using the single sample to define the batch effect, and therefore also the condition effect.
Similar Posts
Loading Similar Posts
Traffic: 824 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thank you, Michael. How? Could you provide more details? Can I just do it as usual, I mean to include batch in the model?
Yes, with the knowledge that, it is expected to be underpowered.
Michael, Thank you very much!