Entering edit mode
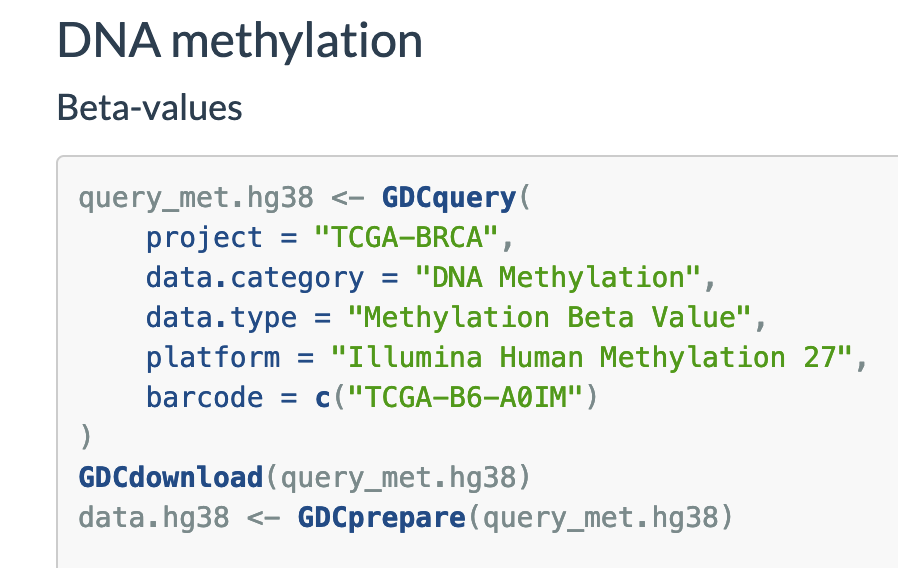
As the code block here , screenshot below.
It worked to get data from platform of Illumina Human Methylation 27 for TCGA-B6-A0IM. But, when I tried to change the platform argument to "Illumina Methylation Epic", it gave me an error as below. I assumed Epic may not be available. Firstly, I thought it may relate to the packages version. But, the error is still there after updating TCGAbiolinks to the lastest one.
So, my question is how should I know the platform availability for TCGA? Thanks!
query_met.hg38 <- GDCquery(
project = "TCGA-BRCA",
data.category = "DNA Methylation",
data.type = "Methylation Beta Value",
platform = "Illumina Methylation Epic",
barcode = c("TCGA-B6-A0IM")
)
# include your problematic code here with any corresponding output
--------------------------------------
o GDCquery: Searching in GDC database
--------------------------------------
Genome of reference: hg38
--------------------------------------------
oo Accessing GDC. This might take a while...
--------------------------------------------
ooo Project: TCGA-BRCA
--------------------
oo Filtering results
--------------------
Error in GDCquery(project = "TCGA-BRCA", data.category = "DNA Methylation", :
Please set a valid platform argument from the list below:
=> Illumina Human Methylation 27
=> Illumina Human Methylation 450
other attached packages:
[1] GenomicRanges_1.56.2 GenomeInfoDb_1.40.1 IRanges_2.38.1 S4Vectors_0.42.1 sesameData_1.22.0 ExperimentHub_2.12.0
[7] AnnotationHub_3.12.0 BiocFileCache_2.12.0 dbplyr_2.5.0 BiocGenerics_0.50.0 TCGAbiolinks_2.32.0
loaded via a namespace (and not attached):
[1] DBI_1.2.3 httr2_1.0.5 biomaRt_2.60.1 rlang_1.1.4
[5] magrittr_2.0.3 matrixStats_1.4.1 compiler_4.4.1 RSQLite_2.3.7
[9] reshape2_1.4.4 png_0.1-8 vctrs_0.6.5 rvest_1.0.4
[13] stringr_1.5.1 pkgconfig_2.0.3 crayon_1.5.3 fastmap_1.2.0
[17] XVector_0.44.0 utf8_1.2.4 tzdb_0.4.0 preprocessCore_1.66.0
[21] UCSC.utils_1.0.0 purrr_1.0.2 bit_4.5.0 xfun_0.48
[25] zlibbioc_1.50.0 cachem_1.1.0 jsonlite_1.8.9 progress_1.2.3
[29] blob_1.2.4 DelayedArray_0.30.1 BiocParallel_1.38.0 parallel_4.4.1
[33] prettyunits_1.2.0 R6_2.5.1 stringi_1.8.4 RColorBrewer_1.1-3
[37] sesame_1.22.2 Rcpp_1.0.13 SummarizedExperiment_1.34.0 knitr_1.48
[41] wheatmap_0.2.0 downloader_0.4 R.utils_2.12.3 readr_2.1.5
[45] Matrix_1.7-0 tidyselect_1.2.1 abind_1.4-8 yaml_2.3.10
[49] codetools_0.2-20 curl_5.2.3 lattice_0.22-6 tibble_3.2.1
[53] plyr_1.8.9 Biobase_2.64.0 withr_3.0.1 KEGGREST_1.44.1
[57] xml2_1.3.6 Biostrings_2.72.1 pillar_1.9.0 BiocManager_1.30.25
[61] filelock_1.0.3 MatrixGenerics_1.16.0 generics_0.1.3 BiocVersion_3.19.1
[65] hms_1.1.3 ggplot2_3.5.1 munsell_0.5.1 scales_1.3.0
[69] glue_1.8.0 tools_4.4.1 data.table_1.16.2 XML_3.99-0.17
[73] grid_4.4.1 tidyr_1.3.1 AnnotationDbi_1.66.0 colorspace_2.1-1
[77] GenomeInfoDbData_1.2.12 cli_3.6.3 rappdirs_0.3.3 fansi_1.0.6
[81] S4Arrays_1.4.1 dplyr_1.1.4 gtable_0.3.5 R.methodsS3_1.8.2
[85] TCGAbiolinksGUI.data_1.24.0 digest_0.6.37 SparseArray_1.4.8 memoise_2.0.1
[89] R.oo_1.26.0 lifecycle_1.0.4 httr_1.4.7 mime_0.12
[93] bit64_4.5.2


Yes, it does. But, I mean how should I know in advance or where can I find the information before running code. Because I have several projects which need downloading. I'd like to avoid the error in my loop. Sorry for the confusion.
I don't see any evidence that TCGA has any EPIC data, but there may be some. Going to their webpage is an exercise in frustration and I didn't try too hard to figure out how it's meant to work. But anyway, you might just add a
tryCatchWE(https://search.r-project.org/CRAN/refmans/simsalapar/html/tryCatchWE.html) to your code to continue in the face of errors.Thanks a lot!
Sorry to get another error.
I want to download it in hg19, I used the code as below. It looks like legacy = TRUE does not work.
Hi @James W. macDonald, I want to check if "unused argument (legacy = TRUE)" due to the GDC legacy archive retirement?
There is a NEWS file for almost all packages that provides, uh, news about the package. You can always read it on the landing page for the package.
Or you can read it directly in R