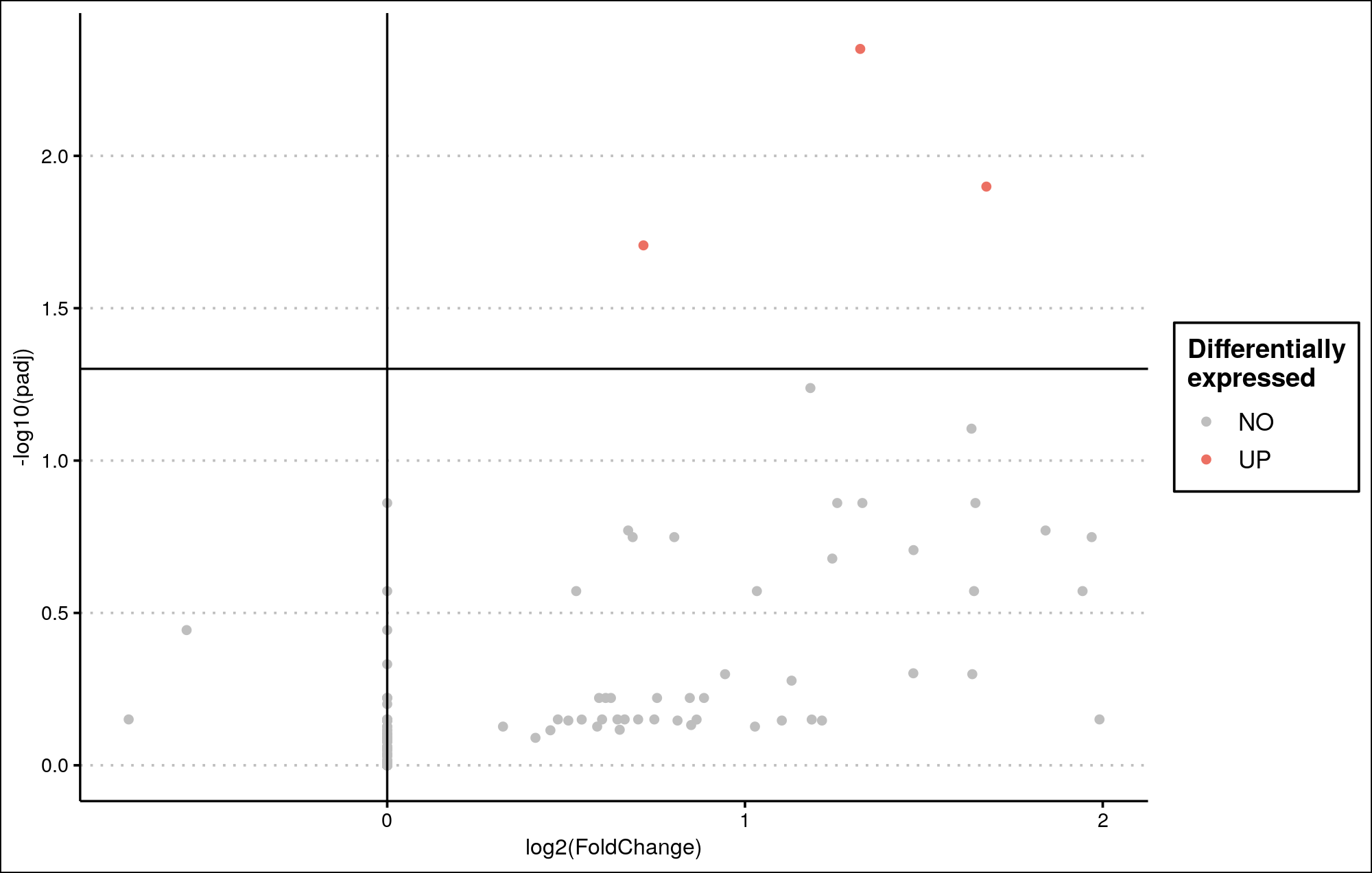
I generated a volcano plot using DESeq2 for a single-cell study transformed into pseudobulk. I am applying log fold change shrinkage using the apeglm method.
However, the resulting plot looks unusual. It does not resemble a typical volcano plot, and many genes have an LFC of 0, yet show varying adjusted p-values (padj).
Are there specific parameters I should adjust when analyzing pseudobulk transformed counts from single-cell RNA-seq?