Enter the body of text here
Hello,
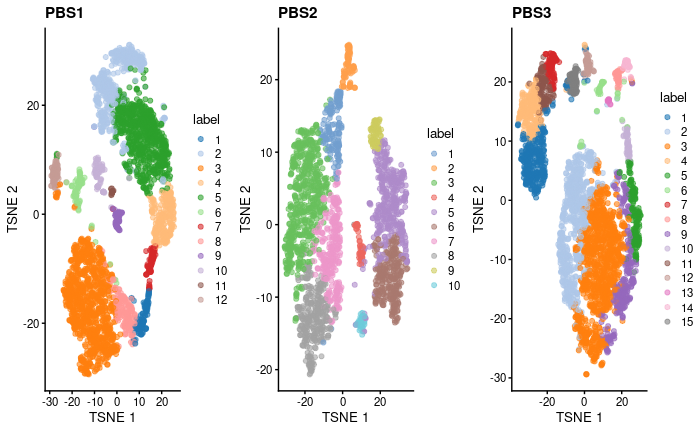
I have 1 "file" that contains scRNAseq data from cells derived from 3 mice, each labelled with one CMO. The three mice same condition (PBS), where labeled (CMO) and pooled and there was one seq run (1batch, am I right?).
I demultiplexed the 10x data using cell ranger.
I'm using bioconductor following this book/guideline: https://bioconductor.org/books/3.15/OSCA.multisample/human-pbmcs-10x-genomics.html#quality-control
My question is: should the tSNE from biological replicates look somehow similar? I'm thinking in shape but also number of clusters. How can I be confident that cluster changes after treatment are due to treatment if there is such "variability" in my PBS triplicates?
I hope the question makes sense
Sorry if this has been questioned before Cheers Jose G


good ideal