Hi all,
I'm not sure if this is appropriate here so I have posted this also on biostars.
This is bulk RNAseq data.
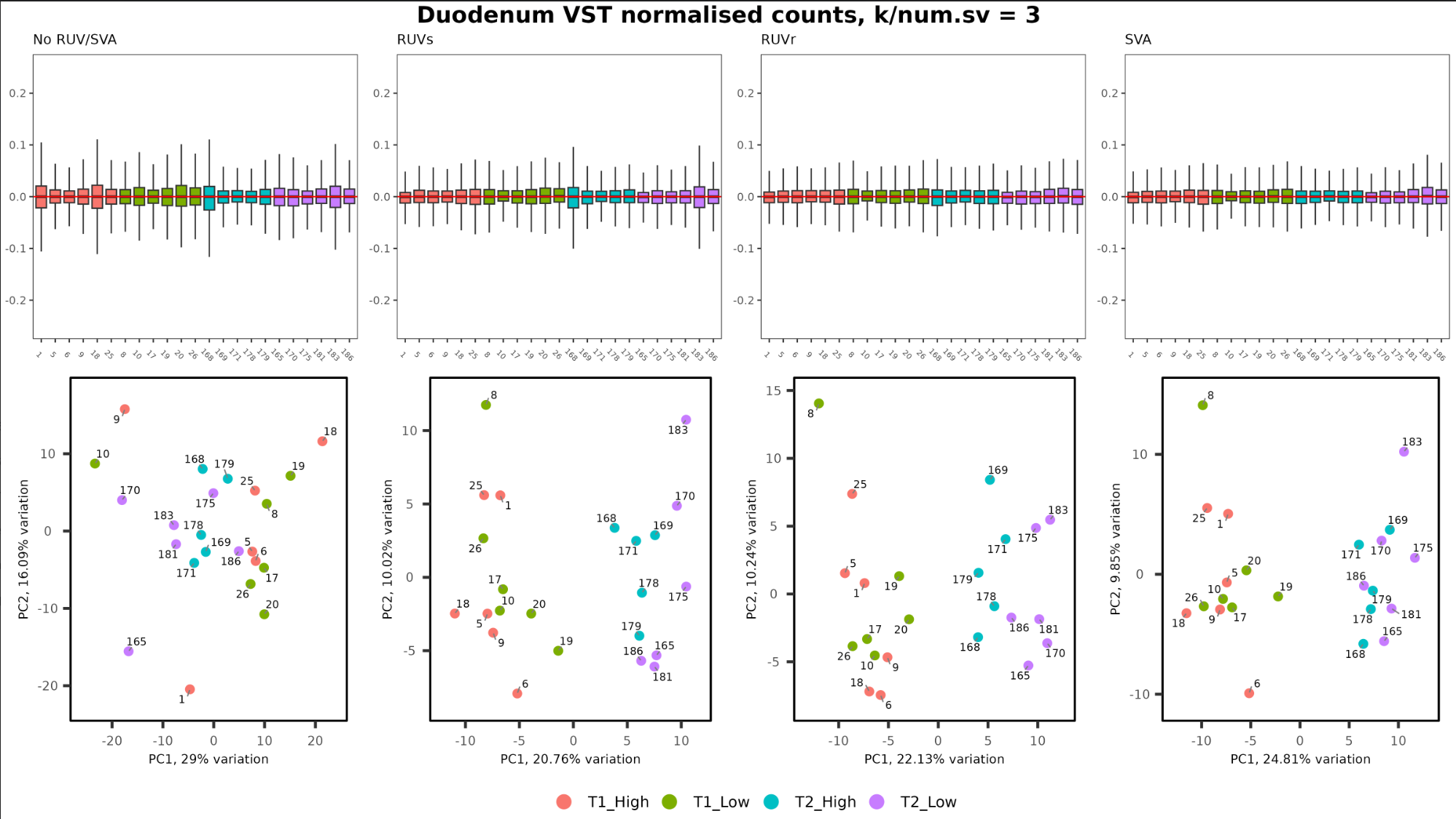
When deciding on the optimal k value for accounting for technical variation, I know I should look for all medians at zero, and box sizes to be equal, but is equality in whisker length between samples a factor in the decision also? I am trying to find the balance between accounting for technical variation, and removing biological signal and fighting the urge to keep increasing k to get better clustering.
In the image below, the left image is the original normalised counts, the others are when using RUVs/RUVr/SVA, where k = 3.
Where k = 1 or 2, a few samples have long whiskers compared to the others (much like in the RUVs RLE plot below), and I don't get separation of clusters on the PCA seen when k = 3.
So I am not sure if:
a) Reasonably equal whisker length is important, go with k = 3
b) Clustering is important, go with k = 3
c) Neither are important, just stop increasing k when the medians are on zero and box sizes are (reasonably) equal, go with k = 1
Thanks All,
Kenneth