Dear all,
I'm a newbie in edgeR and read multiple tutorials + the EdgeR manual but still did not figure out how to solve it.
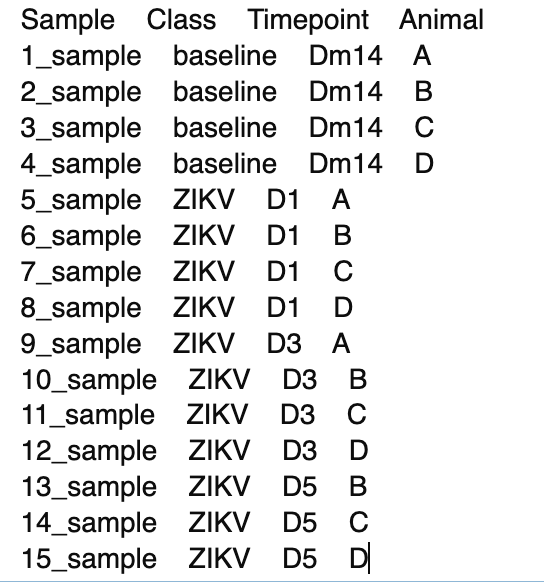
I'm running a DGE analysis with 27 samples. Unfortunately, there are no replicates (yes, I know the problems with this). My data is divided into something like this (a subset of my lib_spec):
This structure follows to 27_sample.
The df is a dataframe output from featureCounts. Columns of samples follow the same order that lib_spec.
Although the obvious comparison would be according to timepoints, I want to run further comparisons between animals, which is the reason why I'm trying to design a complex matrix (following page 42 from edgeR manual).
I tried cbind tp and animal into a column in my lib_spec but it will not help when I run other sorts of comparisons.
My code is:
group <- lib_spec$Class # For creating DGEList group
y <- DGEList(counts = df, group = group) # Creates a table of counts designing each group
bfr_filter <- y$samples # Checks libraries before filtering
keep <- filterByExpr(y) # Creates a vector for genes to be kept based on their expression
y <- y[keep, , keep.lib.sizes = FALSE] # Maintains only what is TRUE in vector list "keep"
aft_filter <- y$samples # Checks libraries after filtering
y <- normLibSizes(object = y, method = "TMM") # Library normalization by a chosen method
group <- lib_spec$Class
tp <- factor(lib_spec$Timepoint) # Covariate 1
animal <- factor(lib_spec$Animal) # Covariate 2
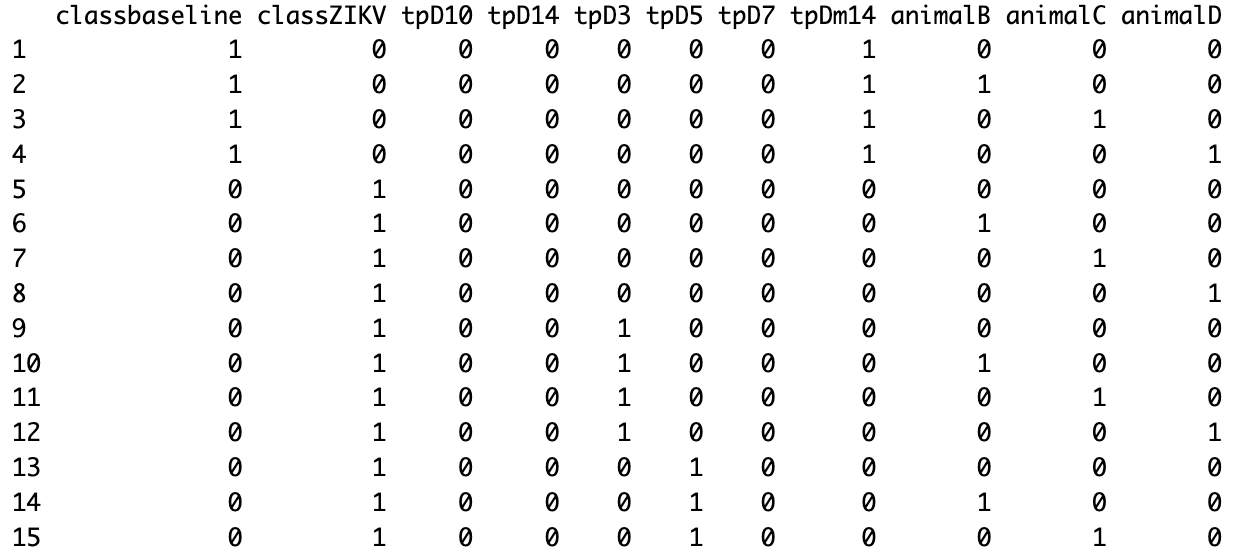
design <- model.matrix(~ 0 + group + tp + animal)
design
However, when I run it, I get the following:
It seems that I "lost" some data on my design because: 1) I don't see tpD1 2) I don't see animalA
This error makes estimateDisp a nightmare. And I don't want to subset my df into possible comparisons (like different timepoints and animals).
Does anyone know why I'm getting this design problem?
In time, does anyone know how to sort design as:
classbaseline classZIKV tpDm14 tpD1 tpD3 tpD5 tpD7 tpD10 tpD14 animalA animalB animalC animalD
Thank you in advance!
Karla