In one article, they used limma to fit a regression between each gene in their mRNA data with a specific protein expression. They report associated genes with that protein as log2FC. here is my question.
- How is limma handling a continuous variable in the model.matrix(lets LKB1 expression)
- How is the logFC calculated for each gene associated with LKB1 expression as LKB1 itself is a continuous variable? Or is it just the estimated cof?
- How it should be interpreted given that I have such data and have output as below
Thank you!!

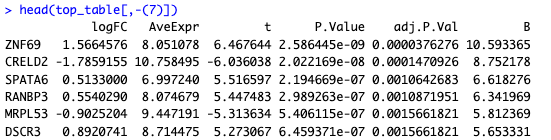

limma fits linear models (as it says on the can!) and the
logFCis just the fitted coefficient. It is exactly the same as for edgeR, which you asked about 4 years ago: Design edger with one or more continues variablesWhat measure of "LKB1 expression" are you entering into the limma linear model? Is it log2CPM or something else? The interpretation of the coefficient obviously depends on what you are entering into the linear model.
The LKB1 expression is in log2 of protein abundance and the gene expression profile is in TPM.
limma is designed to analyse log-expression values. If the data is RNA-seq, then either logCPM or raw read counts input to voom are preferable. I have said before on this forum that I do not consider TPM to be a suitable measure of expression for differential expression analyses. You have attached the "voom" tag to your question, I hope that doesn't mean you are inputing TPM to voom, because you absolutely should not do that.
Anyway, if LKB1 is log2 protein abundance and y was log-expression, then the logFC coefficient in the limma output would be the log2-fold-change in the response gene expression that results from each doubling of LKB1 abundance (as James MacDonald has already said below).