Hi!
I'm trying to plot profile and heatmap for my samples with the following meta data:
> samples
SampleID Tissue Factor Condition Treatment Replicate Control_id
1 HEP152_T5B_1 HEP152_T5B Tumor NAFLD NA 1 HEP152_1_INP
2 HEP152_T5B_3 HEP152_T5B Tumor NAFLD NA 3 HEP152_3_INP
3 HEP152_T5B_4 HEP152_T5B Tumor NAFLD NA 4 HEP152_4_INP
4 HEP262T1 HEP262_T1 Tumor HEP_B NA 1 HEP262T2B_IN
5 HEP262T3 HEP262_T3 Tumor HEP_B NA 1 HEP262T2B_IN
6 HEP268_N1_2 HEP268N1 Normal HEP_B NA 1 HEP268N_INP
7 HEP268_N1_L3 HEP268N1 Normal HEP_B NA 2 HEP268N_INP
8 HEP276_N HEP276_N Normal NAFLD NA 1 HEP276N_INP
9 HEP276N_L1 HEP276_N Normal NAFLD NA 2 HEP276N_INP
I have previously created the following contrasts and performed dba.analyze() for the differential enrichment analysis:
dba.show(dbObj_analyzed, bContrasts = TRUE)
Factor Group Samples Group2 Samples2 DB.DESeq2
1 Factor Tumor 5 Normal 4 5316
2 Condition HEP_B 4 NAFLD 5 5703
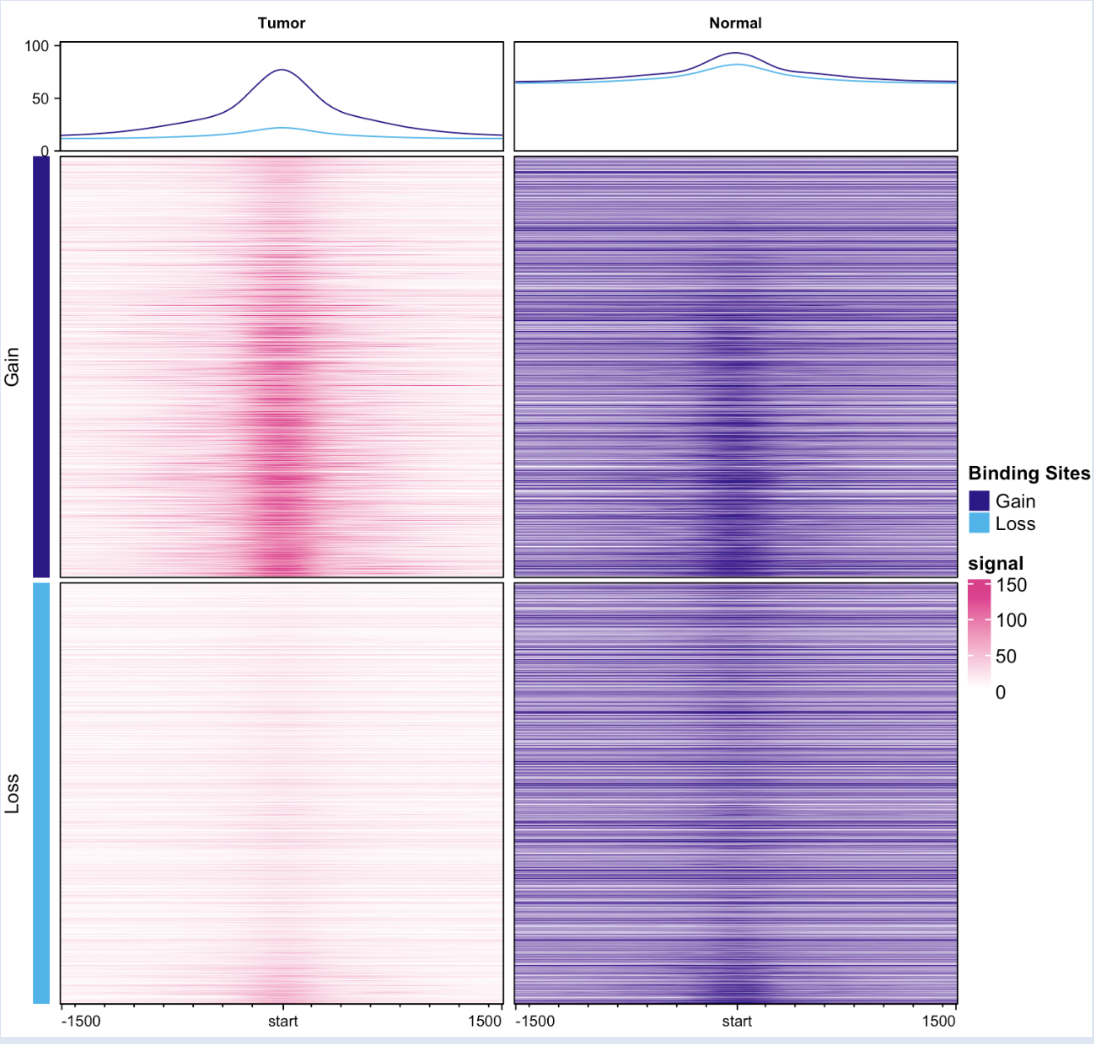
I suppose that if I wanted to present the profile plot and heatmap for tumor vs normal only, I have to merged by replicates, condition (Hep B or NAFLD), and tissue. Therefore, the plots are divided by Factor (Tumor vs normal) only, which turned out to be expected.
profile_merged_TvsN <- dba.plotProfile(dbObj_analyzed,
merge=c(DBA_CONDITION,
DBA_REPLICATE,
DBA_TISSUE))
dba.plotProfile(profile_merged_TvsN)
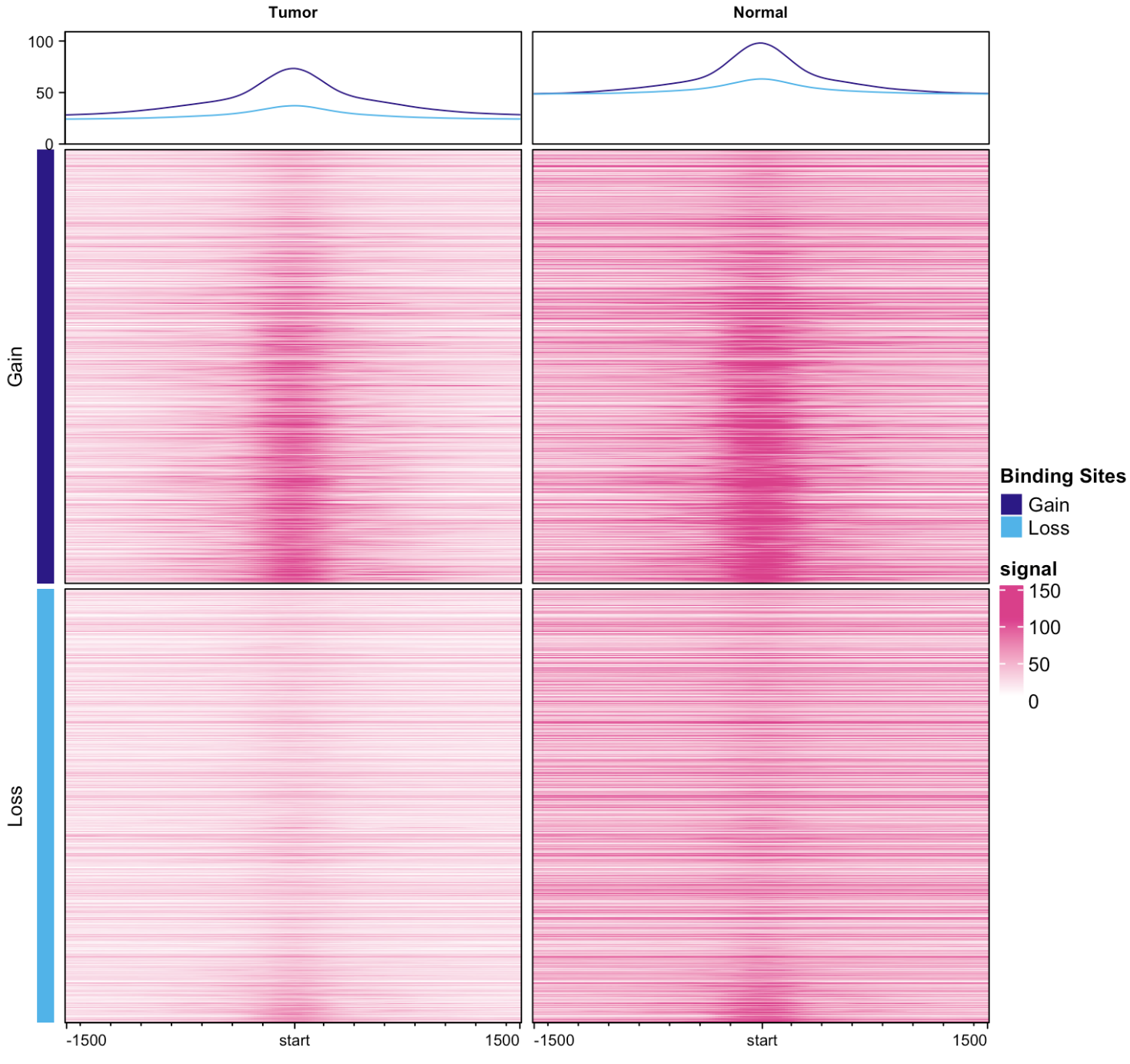
However, when I applied the same logic to present the profile plot and heatmap for HEP_B vs NAFLD only, through merging replicates, factor (tumor or normal), and Tissue, it didn't seem to work as expected.
profile_merged_cond <- dba.plotProfile(dbObj_analyzed,
merge=c(DBA_FACTOR,
DBA_REPLICATE,
DBA_TISSUE))
dba.plotProfile(profile_merged_cond)
May I ask what happened here, and how I can fix this to show the profile plot and heatmap for HEP_B vs NAFLD only?
Also, on a side note, I am curious about the interpretation of my profile plot for Tumor vs Normal. I couldn't decipher the meaning just by reading the diffbind documentation. Here, it seems like the gain curve is above the loss curve for both the tumor and normal. Could anyone break down what each component of the plot and heatmap is saying please?
Thank you very much!
sessionInfo( )
R version 4.3.3 (2024-02-29)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.1.2
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[4] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[7] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[10] tidyverse_2.0.0 DiffBind_3.12.0 SummarizedExperiment_1.32.0
[13] Biobase_2.62.0 MatrixGenerics_1.14.0 matrixStats_1.3.0
[16] GenomicRanges_1.54.1 GenomeInfoDb_1.38.8 IRanges_2.36.0
[19] S4Vectors_0.40.2 BiocGenerics_0.48.1
loaded via a namespace (and not attached):
[1] fs_1.6.4 bitops_1.0-7
[3] enrichplot_1.22.0 doParallel_1.0.17
[5] HDO.db_0.99.1 httr_1.4.7
[7] RColorBrewer_1.1-3 numDeriv_2016.8-1.1
[9] tools_4.3.3 DT_0.33
[11] utf8_1.2.4 R6_2.5.1
[13] lazyeval_0.2.2 GetoptLong_1.0.5
[15] apeglm_1.24.0 withr_3.0.0
[17] prettyunits_1.2.0 gridExtra_2.3
[19] preprocessCore_1.64.0 cli_3.6.2
[21] scatterpie_0.2.2 SQUAREM_2021.1
[23] mvtnorm_1.2-4 mixsqp_0.3-54
[25] Rsamtools_2.18.0 yulab.utils_0.1.4
[27] R.utils_2.12.3 DOSE_3.28.2
[29] plotrix_3.8-4 BSgenome_1.70.2
[31] invgamma_1.1 bbmle_1.0.25.1
[33] limma_3.58.1 rstudioapi_0.16.0
[35] RSQLite_2.3.6 shape_1.4.6.1
[37] generics_0.1.3 gridGraphics_0.5-1
[39] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2 BiocIO_1.12.0
[41] hwriter_1.3.2.1 gtools_3.9.5
[43] GO.db_3.18.0 Matrix_1.6-5
[45] interp_1.1-6 profileplyr_1.18.0
[47] fansi_1.0.6 abind_1.4-5
[49] R.methodsS3_1.8.2 lifecycle_1.0.4
[51] yaml_2.3.8 edgeR_4.0.16
[53] gplots_3.1.3.1 qvalue_2.34.0
[55] SparseArray_1.2.4 BiocFileCache_2.10.2
[57] grid_4.3.3 blob_1.2.4
[59] promises_1.3.0 crayon_1.5.2
[61] bdsmatrix_1.3-7 lattice_0.22-6
[63] cowplot_1.1.3 GenomicFeatures_1.54.4
[65] KEGGREST_1.42.0 magick_2.8.3
[67] ComplexHeatmap_2.18.0 pillar_1.9.0
[69] knitr_1.45 soGGi_1.34.0
[71] fgsea_1.28.0 rjson_0.2.21
[73] boot_1.3-30 systemPipeR_2.8.0
[75] codetools_0.2-20 fastmatch_1.1-4
[77] glue_1.7.0 ShortRead_1.60.0
[79] ggfun_0.1.4 GreyListChIP_1.34.0
[81] data.table_1.15.4 vctrs_0.6.5
[83] png_0.1-8 treeio_1.26.0
[85] org.Mm.eg.db_3.18.0 gtable_0.3.5
[87] amap_0.8-19 chipseq_1.52.0
[89] emdbook_1.3.13 cachem_1.0.8
[91] xfun_0.43 TxDb.Mmusculus.UCSC.mm9.knownGene_3.2.2
[93] S4Arrays_1.2.1 mime_0.12
[95] tidygraph_1.3.1 coda_0.19-4.1
[97] pheatmap_1.0.12 iterators_1.0.14
[99] statmod_1.5.0 interactiveDisplayBase_1.40.0
[101] rGREAT_2.4.0 nlme_3.1-164
[103] ggtree_3.10.1 bit64_4.0.5
[105] progress_1.2.3 filelock_1.0.3
[107] irlba_2.3.5.1 KernSmooth_2.23-22
[109] colorspace_2.1-0 DBI_1.2.2
[111] DESeq2_1.42.1 tidyselect_1.2.1
[113] bit_4.0.5 compiler_4.3.3
[115] curl_5.2.1 xml2_1.3.6
[117] TxDb.Mmusculus.UCSC.mm10.knownGene_3.10.0 DelayedArray_0.28.0
[119] shadowtext_0.1.3 rtracklayer_1.62.0
[121] scales_1.3.0 caTools_1.18.2
[123] ChIPseeker_1.38.0 rappdirs_0.3.3
[125] tiff_0.1-12 digest_0.6.35
[127] rmarkdown_2.26 XVector_0.42.0
[129] htmltools_0.5.8.1 pkgconfig_2.0.3
[131] jpeg_0.1-10 dbplyr_2.5.0
[133] fastmap_1.1.1 GlobalOptions_0.1.2
[135] rlang_1.1.3 htmlwidgets_1.6.4
[137] shiny_1.8.1.1 EnrichedHeatmap_1.32.0
[139] farver_2.1.1 jsonlite_1.8.8
[141] BiocParallel_1.36.0 R.oo_1.26.0
[143] GOSemSim_2.28.1 RCurl_1.98-1.14
[145] magrittr_2.0.3 GenomeInfoDbData_1.2.11
[147] ggplotify_0.1.2 patchwork_1.2.0
[149] munsell_0.5.1 Rcpp_1.0.12
[151] ape_5.8 viridis_0.6.5
[153] stringi_1.8.3 ggraph_2.2.1
[155] zlibbioc_1.48.2 MASS_7.3-60.0.1
[157] org.Hs.eg.db_3.18.0 AnnotationHub_3.10.1
[159] plyr_1.8.9 parallel_4.3.3
[161] ggrepel_0.9.5 deldir_2.0-4
[163] Biostrings_2.70.3 graphlayouts_1.1.1
[165] splines_4.3.3 circlize_0.4.16
[167] hms_1.1.3 locfit_1.5-9.9
[169] igraph_2.0.3 reshape2_1.4.4
[171] biomaRt_2.58.2 BiocVersion_3.18.1
[173] XML_3.99-0.16.1 evaluate_0.23
[175] latticeExtra_0.6-30 BiocManager_1.30.22
[177] foreach_1.5.2 tzdb_0.4.0
[179] tweenr_2.0.3 httpuv_1.6.15
[181] polyclip_1.10-6 clue_0.3-65
[183] ashr_2.2-63 ggforce_0.4.2
[185] xtable_1.8-4 restfulr_0.0.15
[187] tidytree_0.4.6 later_1.3.2
[189] viridisLite_0.4.2 TxDb.Hsapiens.UCSC.hg38.knownGene_3.18.0
[191] truncnorm_1.0-9 aplot_0.2.2
[193] memoise_2.0.1 AnnotationDbi_1.64.1
[195] GenomicAlignments_1.38.2 cluster_2.1.6
[197] timechange_0.3.0


Hello, Did you ever manage to figure this out?