Entering edit mode
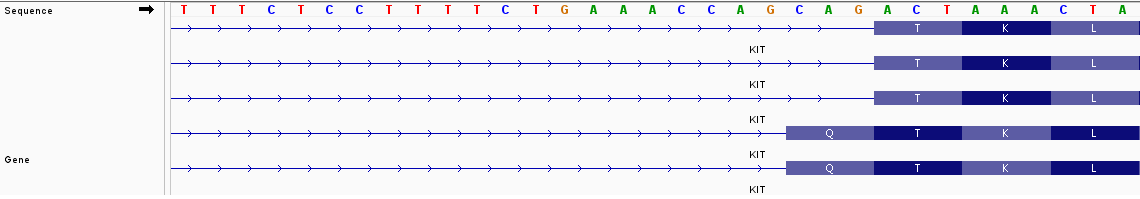
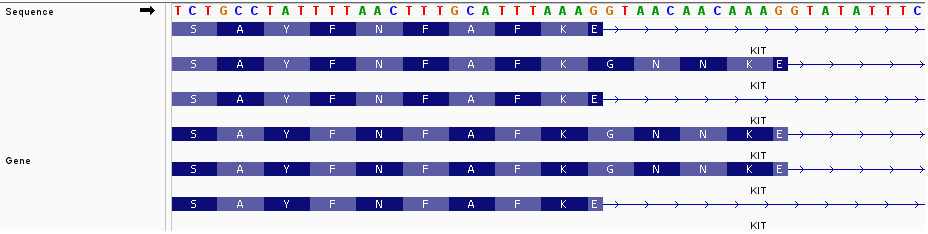
Will Rsubread's Subjunc perform correctly on genes, such as KIT, which have variable-length exons?
I would like to count the reads supporting the Q amino acid exclusion versus Q amino acid inclusion.
Similarly, there is a GNNK inclusion or exclusion elsewhere in the gene.