Hi, Why I am not getting Up and Down regulated genes? My script is as below m I missed something or doing wrong ?
getGEOSuppFiles("GSE18090")
Untar and ReadAffy(rawdata).
gse <- getGEO("GSE18090", GSEMatrix = TRUE)
extracting phenodata and desired column from it as (phenodata$groups).
#Normalization
normdata <- rma(rawdata)
normexprs <- exprs(normdata)
#Limma
design <- model.matrix(~0+phenodata$groups)
colnames(design) <- c("NRM","SVR","CTL")
fit <- lmFit(normexprs, design)
contrasts <- makeContrasts(CTL-NRM, CTL-SVR, NRM-CTL, levels=design)
fit2 <- contrasts.fit(fit, contrasts)
fit2 <- eBayes(fit2)
topTable(fit2)
0
164025
decideTests(fit2)
table(decideTests(fit2))
Results <- decideTests(fit2)
summary(Results)
CTL-NRM CTL-SVR NRM-CTL
Down 0 0 0
NotSig 54675 54675 54675
Up 0 0 0

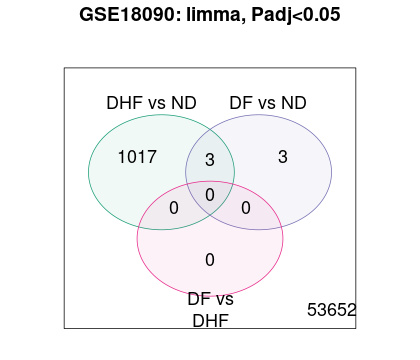

Hi James, Sorry for mistake.
The original paper used individual t-tests, and apparently did not adjust for multiple comparisons. That's not how it's done. In addition, with a t-test you ignore sex-specific differences which is problematic as well. Ideally you would do something like this.
Which provides no evidence for any differential expression. You could try array weights, which helps somewhat, as does relaxing to a 10% FDR