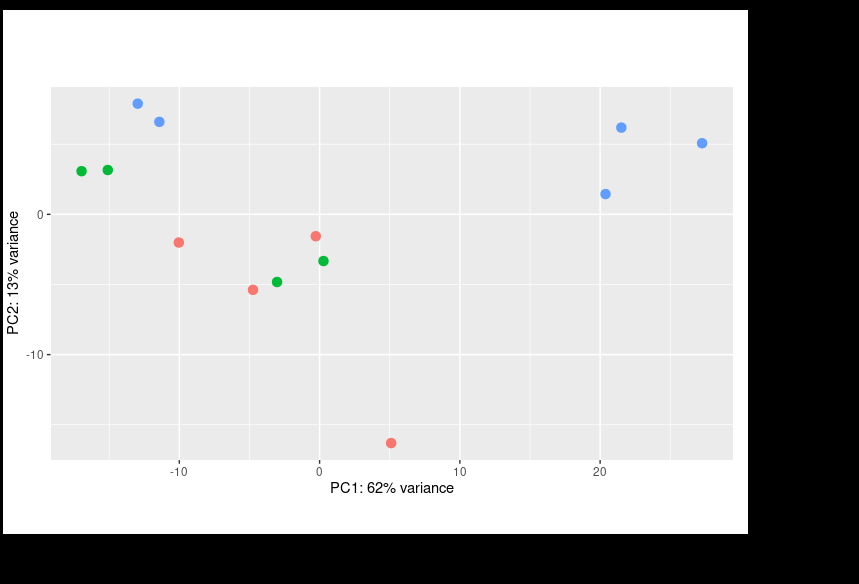
I performed DESEQ on my data implementing the design = ~ condition + batch. I got the result below.
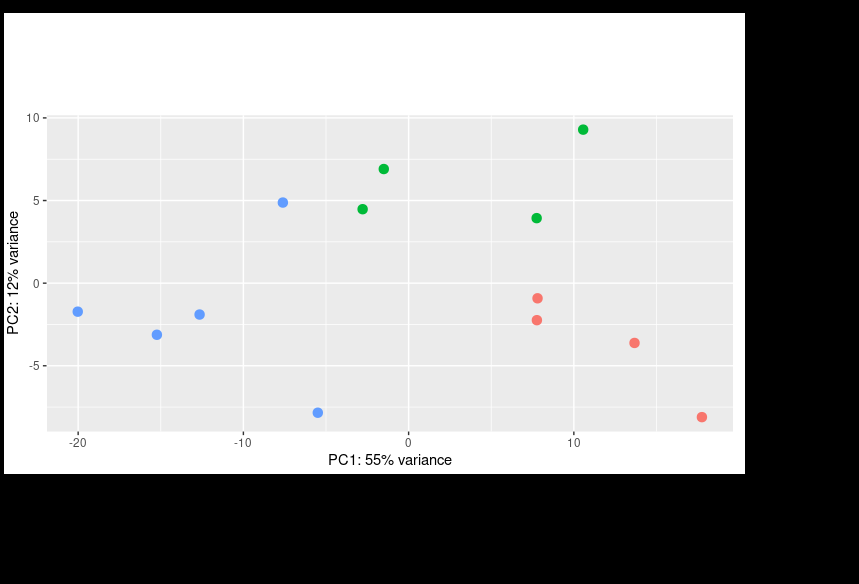
I then corrected the batch effect with Limma and was able to get a better PCA result:
Do I use the Limma batch corrected values to perform my downstream analysis or do I simply proceed with the original DESEQ data? I am reading about it in the forums and it says it is not recommended, but I do not want to proceed to downstream analysis with this batch effect present.
I truly will appreciate any insight. Thank you!