Hi everyone,
I'm running into a problem where the second biological replicates have significantly less IP efficiency and reads than the first replicate, so when I tried running the DESeq2 analysis to find differential enrichment analysis, there were no significant differences between the two dataset even though there are known differences in the literatures.
An example is as such: where the read counts for a certain peak for the first replicate (A_1, B_1) are both higher than the read counts for the second replicate (A_2, B_2) but the over all trend if we are only comparing the replicate itself is that A > B. However because A_2 is lower than B_1 this cause the DESeq2 to not see this as different.
A_1 A_2. B_1 B_2
- 669 756 240
I also tried normalizing the read counts by library sizes, read in peaks but none of them seem to fix the problem as when I did a heatmap or PCA, A_1 will always correspond better to B_1 and A_2 to B_2 because A_1 and B_1 have more reads than A_2 and B_2.
Is there any other way I can approach this problem in terms of normalization? or doing DE on the dataset? Thank you.
```


Okay, thank you, I'll look into it! Also another quick question, when I plotCount for un-normalized counts and normalized counts for a gene that is known to have higher counts in MP than TE, I get the following: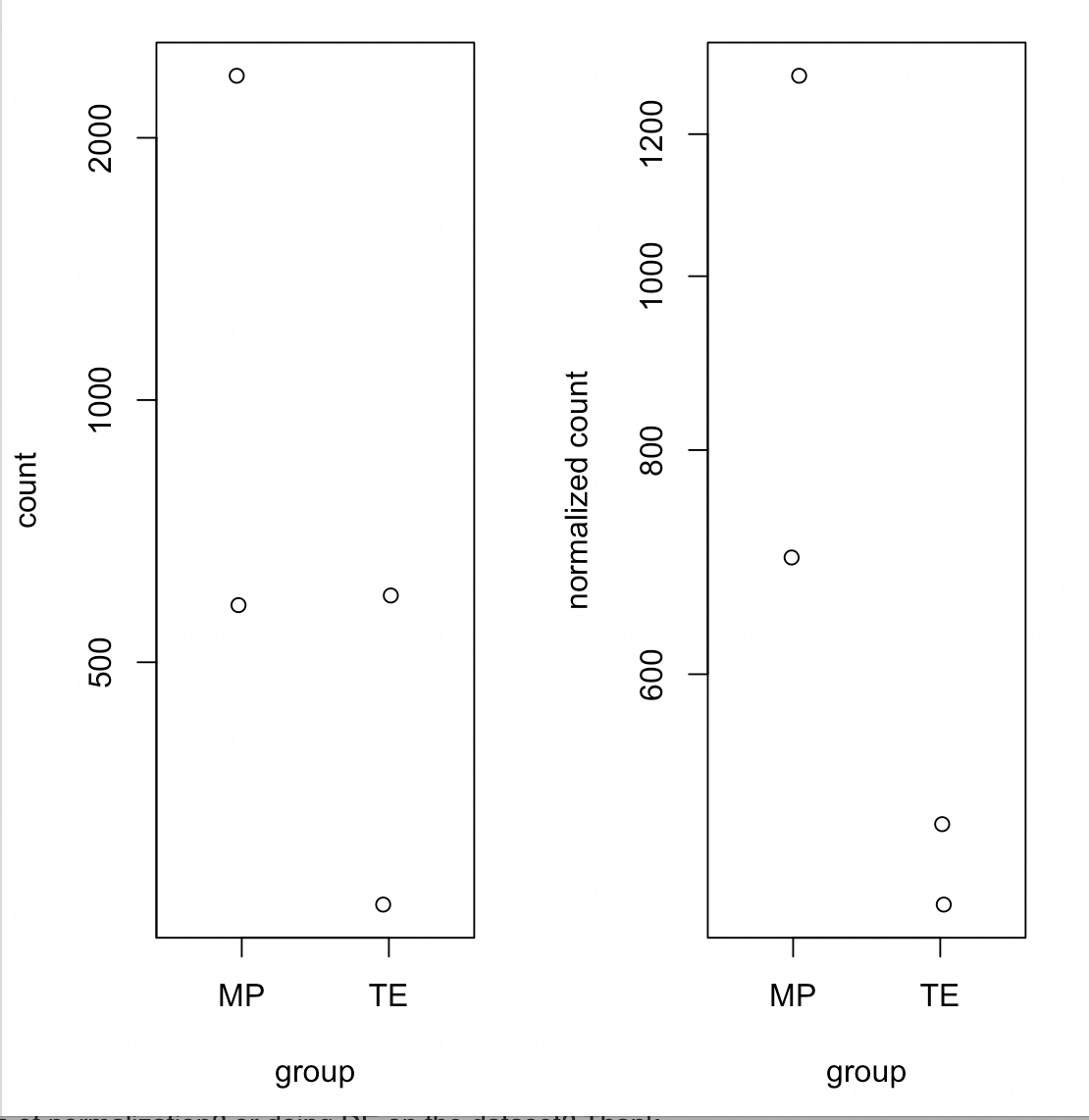 Which the noramlization counts showed the trend expected which is MP > TE, but it is not reported in DESeq2 analysis. Is this because of the huge differences between the replicated in MP?
Which the noramlization counts showed the trend expected which is MP > TE, but it is not reported in DESeq2 analysis. Is this because of the huge differences between the replicated in MP?
Yes, the dispersion factors in. This is likely why it doesn't show up, with only two samples.