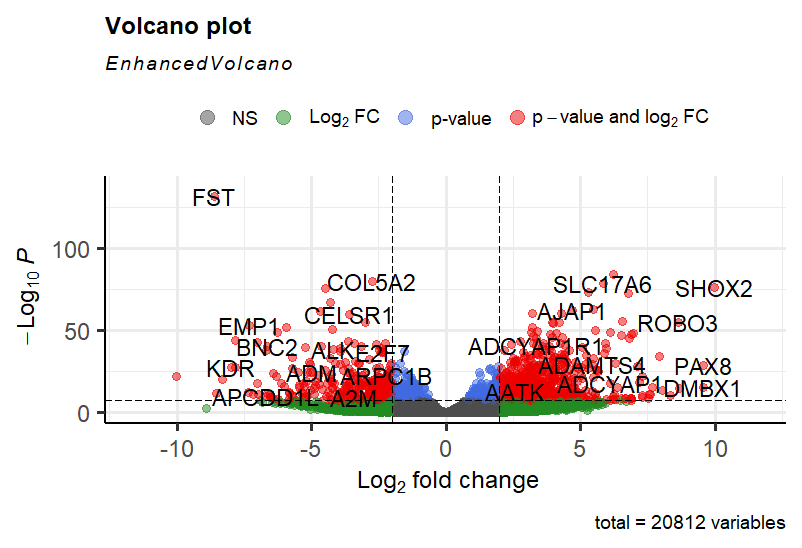
Hi,
I have done a differential expression analysis using DESeq2 and am now trying to visualize my results with a volcano plot. In my plot I would like all DE genes with log2FC >2 and padj < 0.001, and the top 20 genes based on log2FC to be labeled. However, it is not the genes with highest absolute log2FC that get labeled, some genes seem to get labeled based on their lower p-value. Is it possible to adjust the settings for this? Or will I have to label the specific genes using 'selectLab'?
Thanks in advance!
//Sara
EnhancedVolcano(res,
+ lab = rownames(res),
+ x = 'log2FoldChange',
+ y = 'pvalue',
+ title = 'Volcano plot',
+ pCutoff = 6e-8,
+ FCcutoff = 2,
+ pointSize = 3.0,
+ labSize = 6.0)
sessionInfo( )
R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22621)
Matrix products: default
locale:
[1] LC_COLLATE=English_Sweden.utf8 LC_CTYPE=English_Sweden.utf8 LC_MONETARY=English_Sweden.utf8
[4] LC_NUMERIC=C LC_TIME=English_Sweden.utf8
time zone: Europe/Stockholm
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] EnhancedVolcano_1.18.0 dplyr_1.1.2 ggrepel_0.9.4 DESeq2_1.40.2
[5] SummarizedExperiment_1.30.2 Biobase_2.60.0 MatrixGenerics_1.12.2 matrixStats_1.0.0
[9] GenomicRanges_1.52.0 GenomeInfoDb_1.36.1 IRanges_2.34.1 S4Vectors_0.38.1
[13] BiocGenerics_0.46.0 ggplot2_3.4.3
loaded via a namespace (and not attached):
[1] Matrix_1.6-1.1 gtable_0.3.4 compiler_4.3.1 crayon_1.5.2 Rcpp_1.0.11
[6] tidyselect_1.2.0 bitops_1.0-7 parallel_4.3.1 scales_1.2.1 BiocParallel_1.34.2
[11] lattice_0.21-8 R6_2.5.1 XVector_0.40.0 labeling_0.4.3 S4Arrays_1.0.4
[16] generics_0.1.3 tibble_3.2.1 DelayedArray_0.26.6 munsell_0.5.0 GenomeInfoDbData_1.2.10
[21] pillar_1.9.0 rlang_1.1.1 utf8_1.2.3 cli_3.6.1 withr_2.5.1
[26] magrittr_2.0.3 zlibbioc_1.46.0 locfit_1.5-9.8 grid_4.3.1 rstudioapi_0.15.0
[31] lifecycle_1.0.3 vctrs_0.6.3 glue_1.6.2 farver_2.1.1 codetools_0.2-19
[36] RCurl_1.98-1.12 fansi_1.0.4 colorspace_2.1-0 tools_4.3.1 pkgconfig_2.0.3