Good morning everyone,
I have just started working with the SingleR package for annotation of my single cell clusters and was wondering the following:
So far I have only done the annotation for one patient via
#Annotate by patient
pbmc_counts_1 <- GetAssayData(kid.filtered, slot = 'counts.Patient_1')
pred <- SingleR(test = pbmc_counts_1,
ref = ref,
labels = ref$label.main)
However, now I have combined several patients into one Seurat object and I would like to annotate them at the same time.
How can I do this?
Thank you, Bine

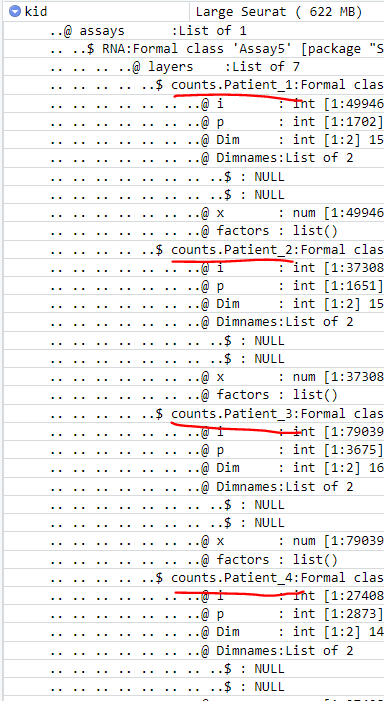

Thank you for your comment & help. When I try to run it with clusters it complains about having multiple layers:
Ah, I just found the function JoinLayers.. I think this should help...