A rather large amount of entries contain a whole lot of genes with identical expression levels, especially some that should be vastly different, i.e. HLA-A and HLA-B.
Here's an example after fetching data using depmap_copyNumber(): depmap_id: ACH-000038 log_copy_number: 1.004462
which is found in 2550 different genes.
Here's another one:
I think I might be missing something here, could anyone help?

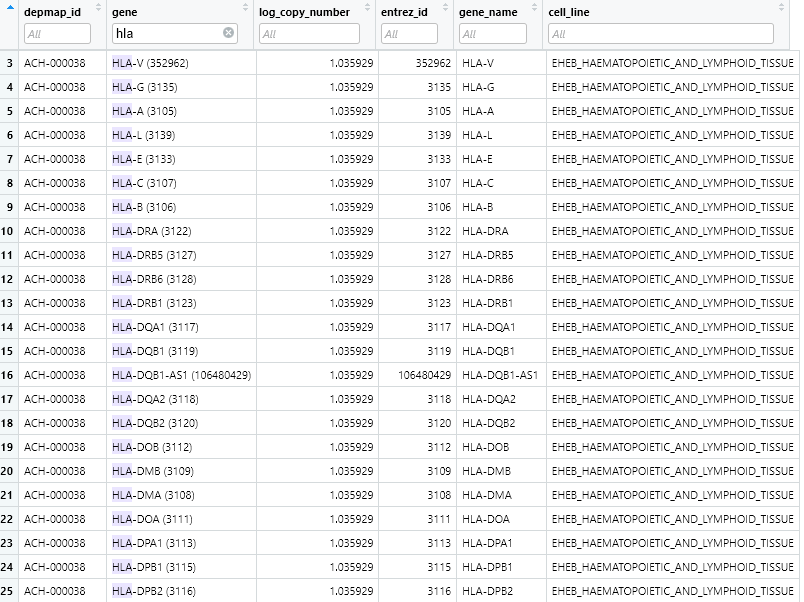

Do you know what copy number means? It's not a measure measure of gene expression