Entering edit mode
Hello! I am trying to use erccdashboard with the option erccmix="Single", since I have only samples with mix1. I'm struggling to understand how to run "runDashboard" in this case. The Vignettes states that I just need to state erccmix="Single". When I run the code with erccmix="RatioPair", everything runs fine. But when I run the same data and inputs with erccmix="Single" I get an error when trying to run "exDat <- dynRangePlot(exDat)":
Error in data.frame(Feature = levels(dataFit$Feature), AveConc = AveConc, :
arguments imply differing number of rows: 74, 73
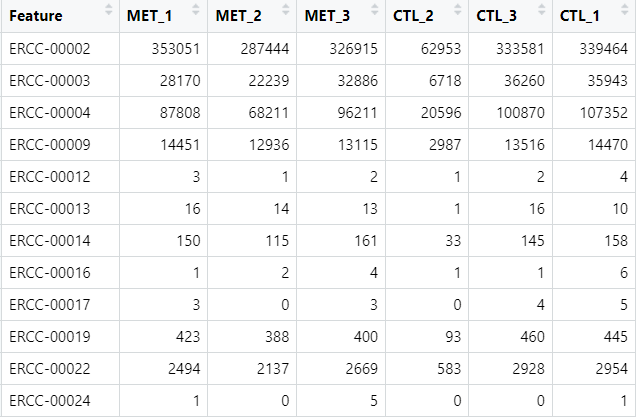
An my data looks like this:
and when I ran this code:
exDat <- initDat(datType="count", isNorm=FALSE,
exTable=data,
filenameRoot="testRun", erccmix="Single",sample1Name="MET",
sample2Name="CTL",
erccdilution=1/40,
spikeVol=1.3, # ul
totalRNAmass=1, # ug
choseFDR=0.1)
exDat <- dynRangePlot(exDat)
The output looks like this:
exDat <- dynRangePlot(exDat)
mRNA fraction estimate is assumed to be equal to 1
Number of ERCCs in Mix 1 dyn range: 85
Number of ERCCs in Mix 2 dyn range: 85
These ERCCs were not included in the signal-abundance plot,
because not enough non-zero replicate measurements of these
controls were obtained for both samples:
ERCC-00017 ERCC-00024 ERCC-00147 ERCC-00061 ERCC-00075
ERCC-00081 ERCC-00083 ERCC-00117 ERCC-00137 ERCC-00138
ERCC-00168
Error in data.frame(Feature = levels(dataFit$Feature), AveConc = AveConc, :
arguments imply differing number of rows: 74, 73