Hi,
I am not sure if this is the expected function behavior or if it is a possible bug.
I have very deep RNA seq data.
I am trying to plot the pileup,
I observe peaks at the start of some exons,
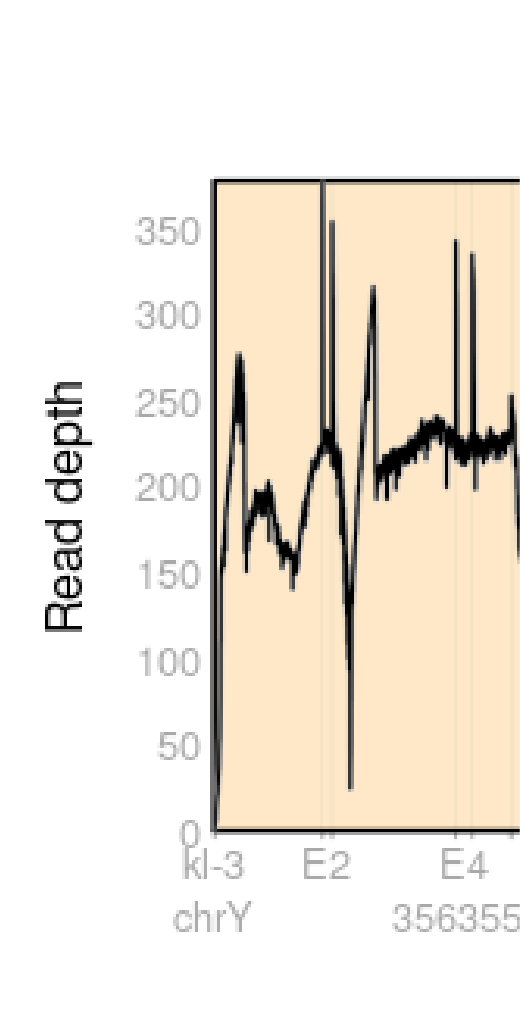
But when I change the max_depth option to my max depth the peaks disappear, and the read depth is precisely the same as in IGV.
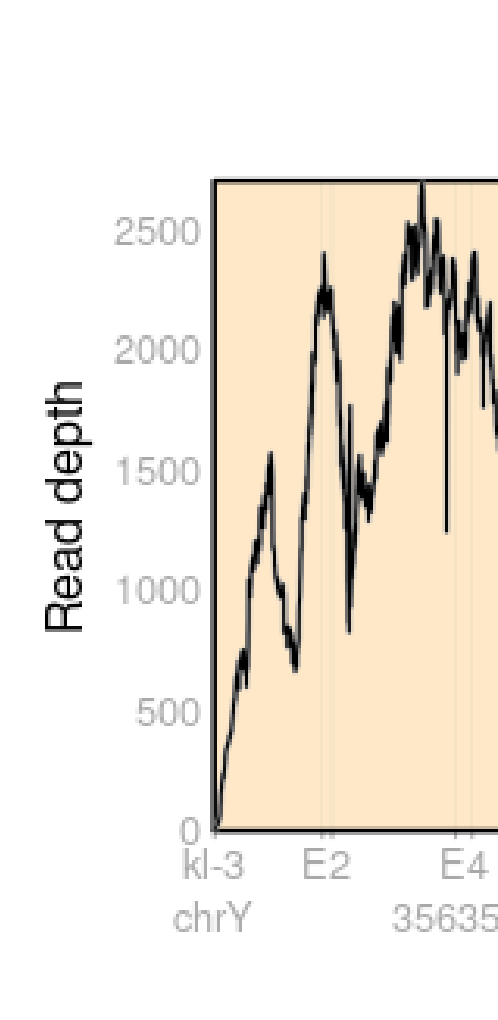
From the documentation, max_depth only count max_depth overlapping reads.
But if so :
1- why are those peaks?
2- in my case, why is not everything max out?
I may have a confusion about what is called overlapping reads...
The only thing I changed between those two plots is the max_depth option.
I greatly appreciate your help,

