Hi all,
I apologize in advance that this is a heavily stats based question but also a plea for guidance on if I am using the right packages currently.
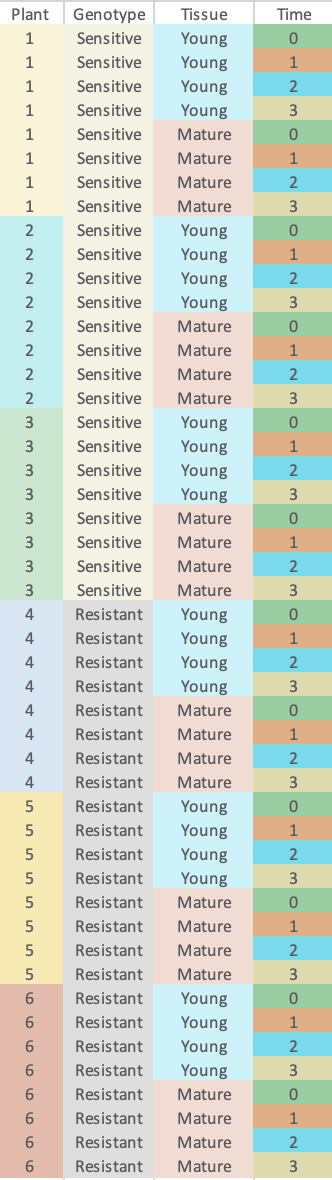
As you can see in the image my RNAseq dataset consists of 6 individual plants, 3 bio-reps of one genotype, and three of another. For each of those I have young and mature tissue, a before treatment timepoint followed by 3 post treatment timepoints (15, 60, 180 minutes)
My hypotheses involve identifying differences in expression in response to treatment, related to both genotype and tissue (the resistant mature leaves show a rapid response), but also in just genotype (the response doesn't always segregate with resistance so not convinced it's causal so much as linked, but could be contributing). So I want to compare each Genotype+Tissue at each time point to itself before treatment, then do the same for just genotype, perhaps averaging over tissue.
The only time I am very interested in a direct comparison between Sensitive and Resistant is before treatment for constitutive differences. For the other contrasts I am happy with casually comparing the results of within genotype contrasts (but open to suggestions about the most powerful way to make any of these comparisons)
So it seems to me that each individual does have multiple measurements, but given my experimental questions do I need to include individual/plant as a random effect to control for correlation of each across time, or maybe across tissue? I have been using both DESeq2 and EdgeR just using a full model of genotype + tissue + time and their interactions, I have played around with the portions of the vignettes discussing both within and between comparisons, but I have become very confused.
Should I migrate to Dream to run the proper repeated measures design or is okay to make my contrasts as I am without major increased risk of type I error? If so, I do treat my biological reps as individuals?
I plan to have a stats consult at my university but won't be able to for a couple of weeks, so any help is very much appreciated. .
Thanks!
# include your problematic code here with any corresponding output
# please also include the results of running the following in an R session
sessionInfo( )


Gordon,
Thanks for taking the time to read and reply.
The limma-voom approach seems to have worked great for the duplicateCorrelation implementation. With just a few painless edits to my edgeR script I was able to run all my contrasts and save the output tables. I am intrigued to compare to my initial results, and relieved to move forward with my analysis being a bit more accurate. Looking forward to citing limma.
-Crystal