I have two conditions: negative control and treatment. Each one has 6 replicates. Then, from PCA analysis and heatmap, I found out that one negative control replicate is very different from the rest of the samples and that it is an outlier, so I decided to remove that sample from the dataset for downstream analysis. I proceeded to use fishpond by calling scaleInfReps(), labelKeep(), and swish(). However, swish() returned the following error.
Error in infRepsArray[, cond2, ] : subscript out of bounds
I suspect that this has to do with me having 5 samples in one condition and 6 samples in the other. Is that right? What should I do here?
EDIT: Fishpond version is 2.6.0
EDIT2: These replicates are biological, rather than technical.
EDIT3: I will include the PCA plot below

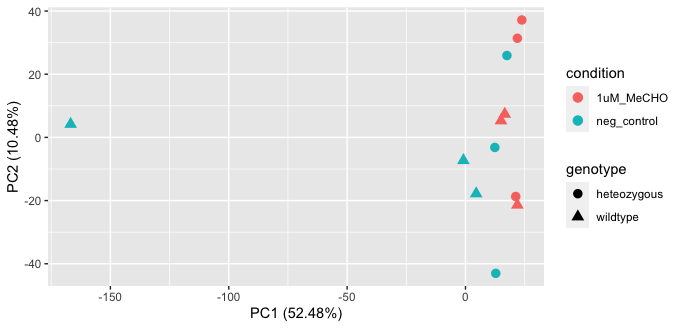

After some debugging, I believe that there is a bug in fishpond codebase. I've opened an issue.