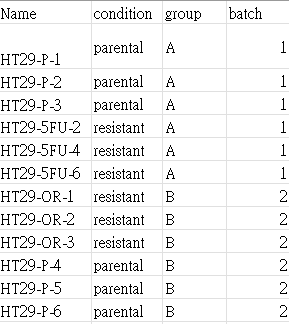
Hi, I'm new in biological analysis.
I want to use DESeq2 to do my analysis, I have batch 1 and batch 2, and the batch equals group.
If I want to remove the batch effect, should I only use design = ~ group + condition, or I should use design = ~ batch + group + condition ?
And this is PCA result, is there any batch effect exist? Should I remove batch effect before WGCNA?

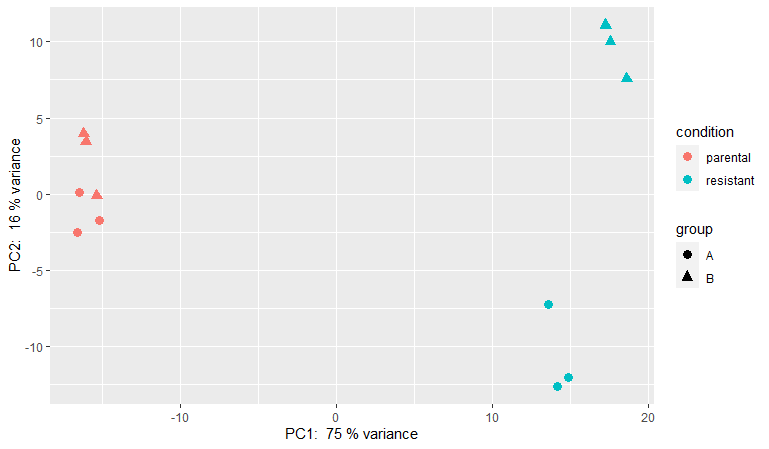

DESeq2 remove batch effect is always my favorite and finally I got the detail here to know how to do it. To get the Brisbane Tree Removal we can find the best services that are good to provide us the best services for tree removal.