Entering edit mode
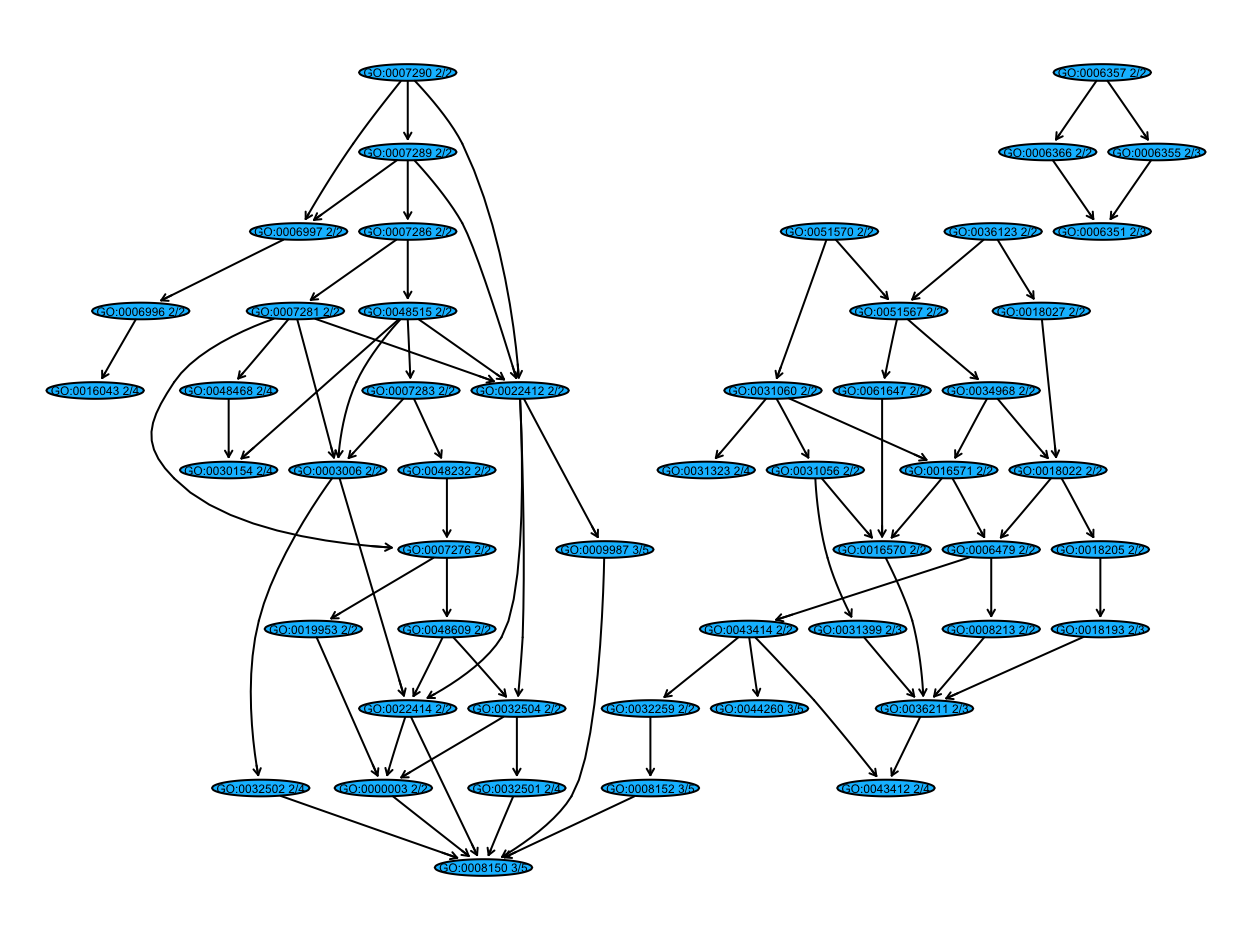
When I used plotGOTermGraph function to make the GO graph, I notice some of the subtrees are not connected to the root node (e.g. GO:0008150 biological process). I wonder if there is a way to connect these subtrees to the root node manually. Or could you recommend any other packages/functions to plot the diagram? Thank you!
I made a small demo below:
entrezUniverse_example <- c("6181", "2915", "8846", "55818", "63946")
selectedEntrez_example <- c("6181", "55818", "63946")
# hypergeometric test
hgCutoff <- 0.001
params_example <- new("GOHyperGParams",
geneIds=selectedEntrez_example,
universeGeneIds=entrezUniverse_example,
annotation="org.Hs.eg.db",
ontology="BP",
pvalueCutoff=hgCutoff,
conditional=FALSE,
testDirection="over")
# perform the test
hgOver_example <- hyperGTest(params_example)
# plot
graph_example <- inducedTermGraph(hgOver_example, id = c(summary(hgOver_example, pvalue = 1)$GOBPID),
children = F, parents = T)
plotGOTermGraph(graph_example, r = hgOver_example,
add.counts = T, max.nchar = 200,
node.colors = c(sig="darkorange", not="deepskyblue1"),
node.shape = "ellipse")
With pvalue = 1, I can plot the full tree and all nodes will be connected to the root node eventually, but this has too many nodes.
graph_example <- inducedTermGraph(hgOver_example, id = c(summary(hgOver_example, pvalue = 0.4)$GOBPID),
children = F, parents = T)
With pvalue = 0.4, I have a subset of nodes but some subtrees are not connected to the root node.