Hi
I have a phenotype of my interest (T versus C). My samples have been collected in several runs(batches).
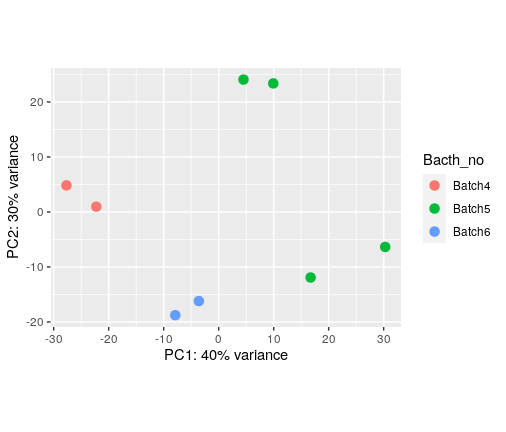
PCA shows that runs can separate my samples
This is how my colData looks
> pData3[,c(1,2,8)]
Bacth_no Samp_no StatuS
s1 Batch4 36 T
s2 Batch4 37 T
s3 Batch5 43 C
s4 Batch5 44 C
s5 Batch5 47 C
s6 Batch5 48 C
s7 Batch6 53 C
s8 Batch6 54 C
>
I am trying these to remove the run effects but I get error
> dds <- DESeqDataSetFromMatrix(countData = exprs_df[ , rownames(pData3)],
+ colData = pData3,
+ design = ~ Bacth_no+Status)
Error in checkFullRank(modelMatrix) :
the model matrix is not full rank, so the model cannot be fit as specified.
One or more variables or interaction terms in the design formula are linear
combinations of the others and must be removed.
Please read the vignette section 'Model matrix not full rank':
vignette('DESeq2')
In addition: Warning message:
In DESeqDataSet(se, design = design, ignoreRank) :
some variables in design formula are characters, converting to factors
> dds <- DESeqDataSetFromMatrix(countData = exprs_df[ , rownames(pData3)],
+ colData = pData3,
+ design = ~ Bacth_no+Status+Bacth_no:Status)
Error in checkFullRank(modelMatrix) :
the model matrix is not full rank, so the model cannot be fit as specified.
One or more variables or interaction terms in the design formula are linear
combinations of the others and must be removed.
Please read the vignette section 'Model matrix not full rank':
vignette('DESeq2')
In addition: Warning message:
In DESeqDataSet(se, design = design, ignoreRank) :
some variables in design formula are characters, converting to factors
>
Please could you tell me a helpful design?
Thanks a million


Thank you very much