Dear Community,
I'm running an ALDEx2 analysis in R on microbiome data and I'm currently wondering if I'm interpreting the data correctly.
code I run for the AldeX2 analysis:
aldexmodel <- aldex(Aldex_mat_fungi_antago, Aldex_variable_fungi_location_antago, mc.samples=128, test="t", effect=TRUE,include.sample.summary=TRUE , denom="all", verbose=FALSE)
The 'treatment' to be tested is "location" which consists of 2 possiblities: "front" or "back".
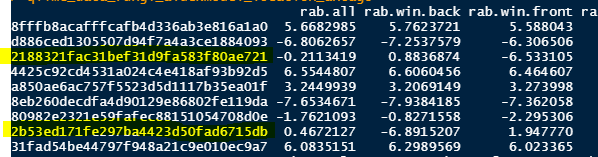
In the model output I can see that I have 2 Features/ASVs with a P-value <0.05 according to the Welsh t-test (we.ep, in output).
I know which 2 features this are, but I want to know if they are more present in the "back" or "front". Therefore I think I need to check the "rab.win" values, shown in the image below (the 2 features are highlighted in yellow):
feature 1 (ending with ...721) has a rab.win value of 0.88 for the 'back' and -6.53 for the front. Do I interpret correctly that this feature is more abundant in "back"
feature 2 (ending iwth ....15db) has a rab.win.back value of -6.9 and a rab.win.front value of 1.94. Do I interpret correctly that this feature is more abundant in "front"?
So the higher the 'rab.win.back' value, the higher the presence of that feature in the treatment?
Thank you in advance.
RvH


I am very impressed with your article. Like you said when you delete a result, run 3 it will be pushed from the left participant column to the right result column. I like it very much.
I value your insight and reaction. Would the advice still apply if I attempted to copy a picture and then paste it? You worked really hard to provide us with engaging the backrooms content.