Hello Everyone,
I am working on a project with the goal of improving the prediction accuracy of melanoma relapse by combining the RNA signature with the clinical features.
Firstly, I calculated the asssociation/relationship between the clinical features with the outcome (early recurrence < 3yrs vs no relapse with at least 5 yrs follow up) and found that Tcat, mitosis and last_vitalstatus are closely related with the outcome result.
Secondly,I need to do the DGE by using the DESeq2 to extract a signature. During this process, I set the closely related clinical features as controls in the design to exclude their effect on the DGE result.
The R code can run successfully, but most of the generated volcano plot are weird when I consider some control factors. Only when I use the last_vitalstatus as control, the volcano plot looks normal (Fig3). The more control factors, the more stretched of the volcano plot, as presented by the last figure(VolcanoPlot_Mitoses_LastVitalStatus_Tcat_Ascontrol.jpeg). If I don't consider any control, the volcano plot is perfect, as presented by the Fig1 (VolcanoPlot_NoControl.jpeg).
The code is attached. I am very grateful, if someone can tell me if I did something wrong. As the Outcome is the feature that I am interested, so I put it at the end of the design.
<h6>#</h6>dds <- DESeqDataSetFromMatrix(countData=RawCountMatrix, colData=EventControl, design= ~ mitoses+Last_vitalstatus+Tcat+Outcome_Grouping)
keep <- rowSums(counts(dds)) >= 10 dds <- dds[keep,]
dds <- DESeq(dds) res <- results(dds)
resOrdered <- res[order(res$padj),] write.csv(as.data.frame(resOrdered), file="Outcome_Good-Bad.csv")
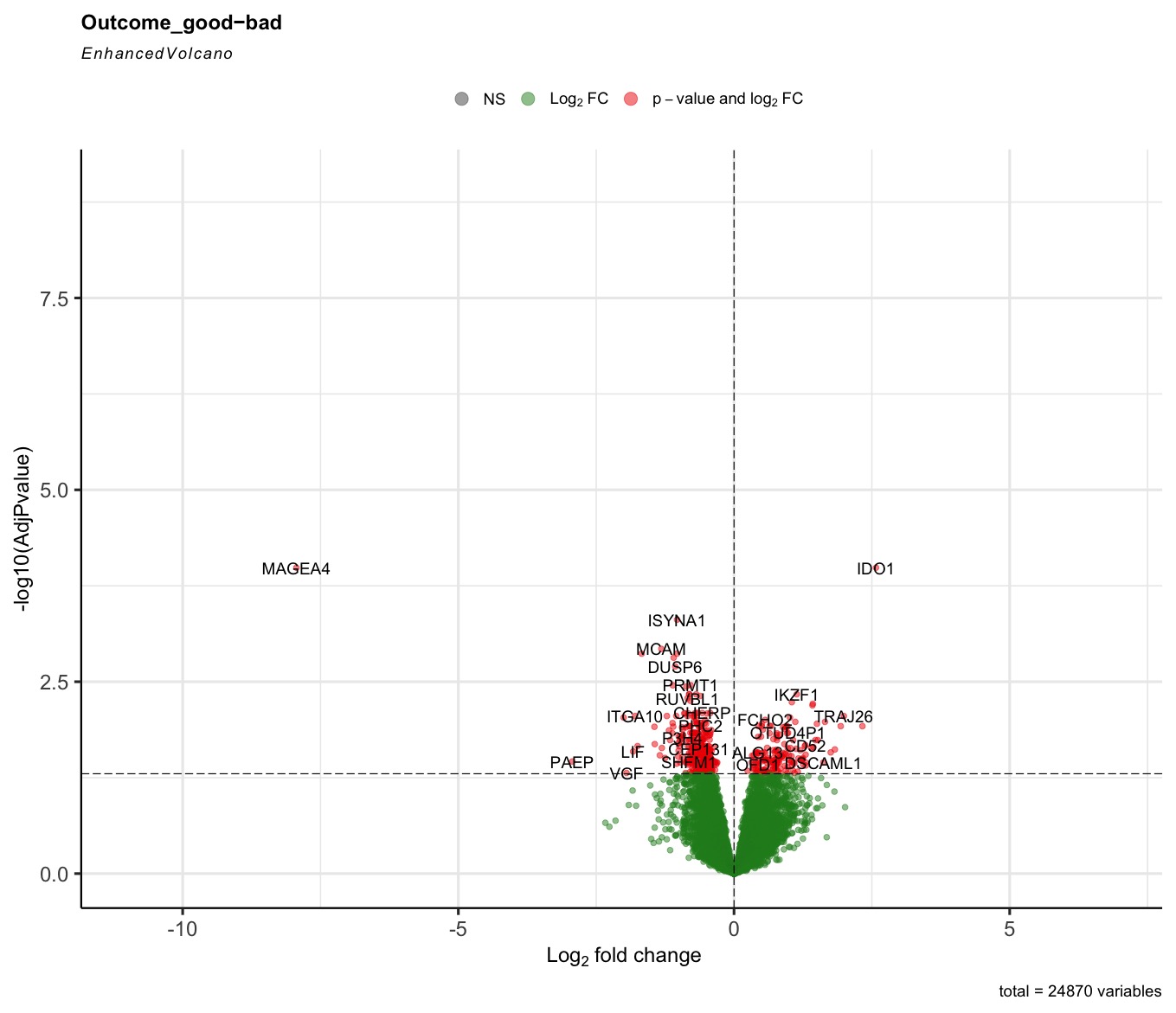
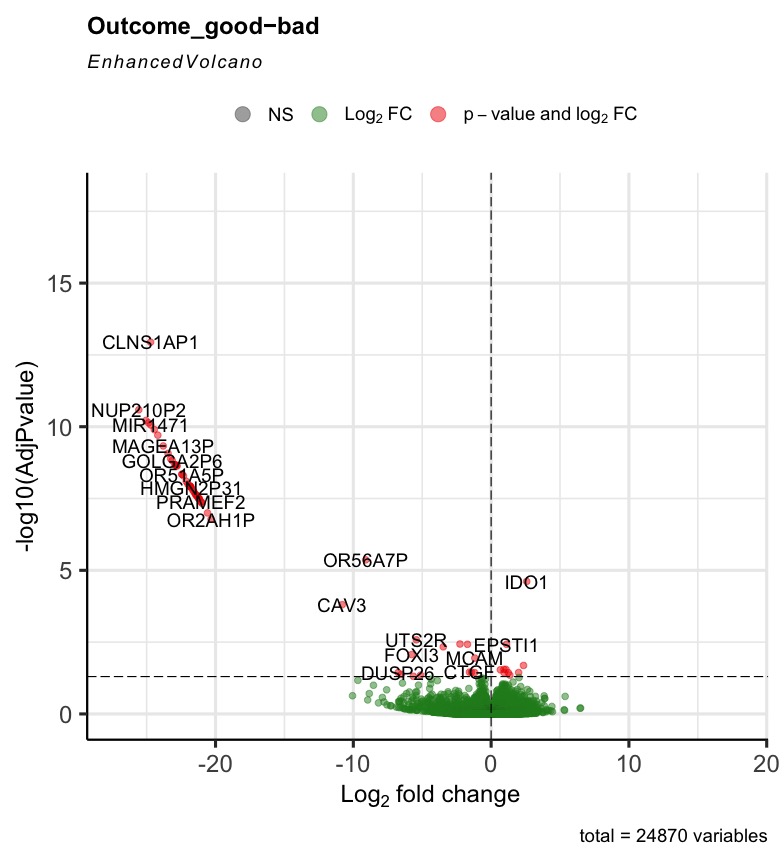
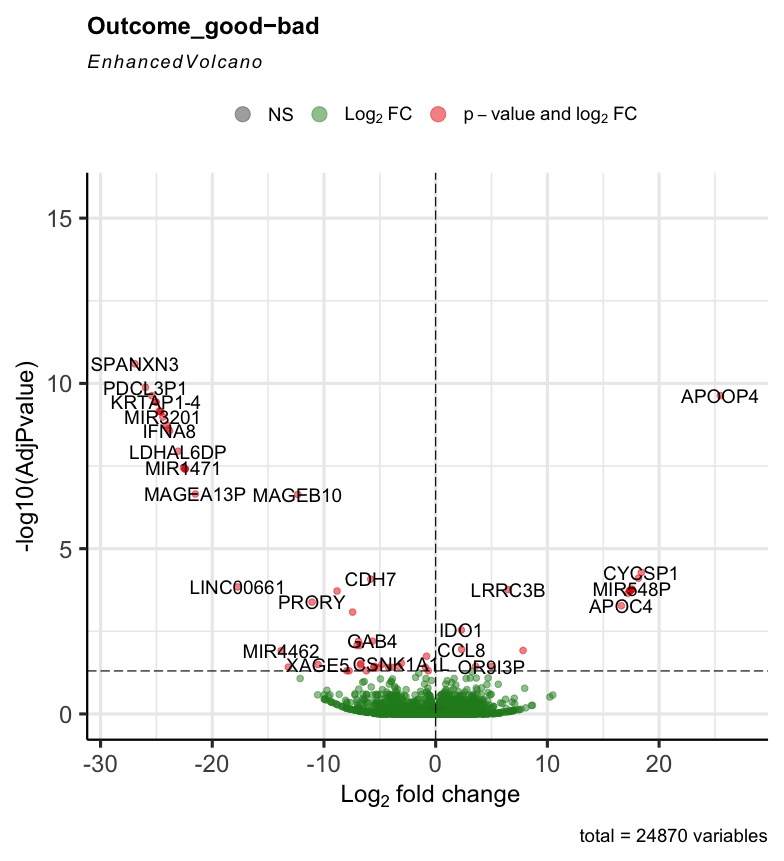
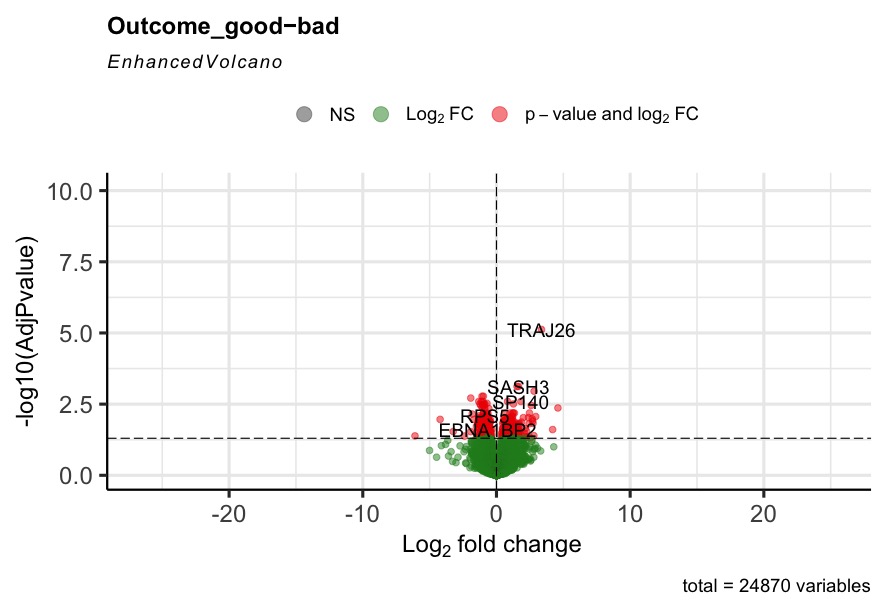


Prefiltering could help. Your filter only removes genes that are not expressed rather than genes with single or few high outliers or inconsistent detection. See the vignette on prefiltering or simply use filterByExpr from edgeR.
Thank you for your response. I will filter the data first to see what is the effect of expression outlier on the result. Once I get result, I will post it here.
Thanks again.