I am Edmond Geraud, a PhD student from the (Technical) University of Valencia (Spain). I am studying a multi-omic integration for a certain syndrome, with several omics, which include metabolomics and metagenomics. The independent variables are factors like sex and obesity.
In order to do a proper integration I need to normalize the sets. For that purpose, I am using the voom function from the limma package. I am aware that it was built under the assumption of continuous data from microarray data.
So, before applying voom I am following the best practices for the metagenomic data obtaining a continuous distribution. Thus, when I use voom to either data set, I get the "log expression values" and some weights according to the design.
The problem is that I do not get the expected results confirmed by previous papers. But the information is there, in other words, given the fact that previous results are accepted, if a do a transformation and a normalization via voom the same information should be there
So what I have done, is the following, assuming X is the data set and the design is the model.matrix object with a certain factor like sex,
v <- voom(X,design=design)
y <- v$E/v$weights.
OR
y <- v$E*v$weights.
In other words, I divided (or multplided, I tried both approximations) the "expression values" by their weights in a pair-wise manner. When I do that I obtain the expected results ( in a PCA), so is this approximation be suitable to consider the weights generated in voom function?
I mean, I know that weights are used for the correction of heteroscedasticity for later calculations in limma such as in weighted linear regression, but my purpose is only to get the normalized data for a later integration,
Does what I am doing make sense? i.e, weighting the "expression values" in that way? Or should I do it in another way? The question is, am I forcing the dataset to get the results with this approximation ?
(Here I attach two PCAs as an example, "PCA_weigths_voom.png" is the output of doing a PCA via v$E/v$weights, the other one, "PCA_without_weights" is the output of doing PCA only with v$E)
Greetings,

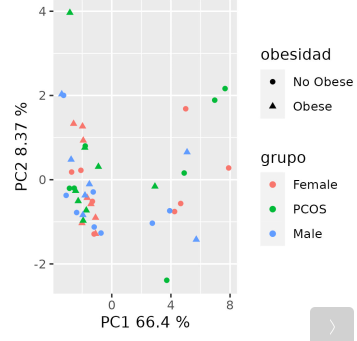
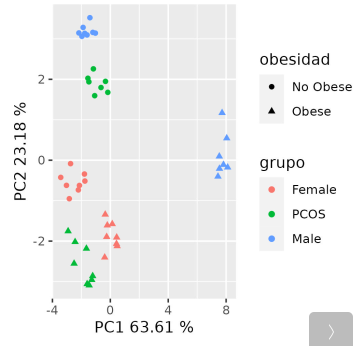

Thank you !
So, even if I remove the log with an exponent, it does not make sense? neither for metabolomic data?
No, precision weights are just not something that you would multiply or divide an expression estimate by. The resulting quantities wouldn't have any meaning. Precision weights are not used for normalization.