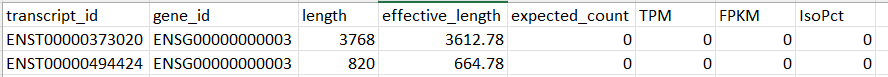Hello,
I have a dataset contains the following columns: transcript_id, gene_id, length, effective_length, expected_count, TPM, FPKM, and IsoPct. And for the same gene_id more than two transript_id .. so it's a transcript-level dataset
I thought I need to use tximport to convert it to counts so I could use it for DESeq2, but I didn't really get how can I convert it exactly.