Entering edit mode
I am working with a 10x Visium dataset and I would like to calculate the Number of Cells per spot in my dataset. I am following the OSTA book at the section linked below. When I run addPerCellQC() it does calculate QC metrics, but 'cell_count' does not seem to be one of them and I am not sure why? The scuttle package seems to have a similar function (perCellQCMetrics) but that does not seem to be producing the desired result either.
Also, is there any notes on what exactly each column of colData is showing? e.g. sum
https://lmweber.org/OSTA-book/quality-control.html#number-of-cells-per-spot

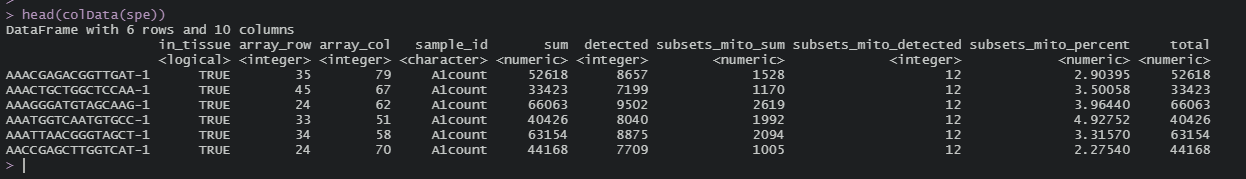

Of the columns there, sum and total both refer to the total number of transcripts per observation (per spot, presumably, for Visium data). Detected is the number of transcripts/genes with non-zero entries for that observation. The subsets_mito sum and detected and similar measures for the mitochondrial gene annotation passed in to the QC function, and subsets_mito_percent is the mito sum as a fraction of the overall sum
Can't help you with the cell count unfortunately