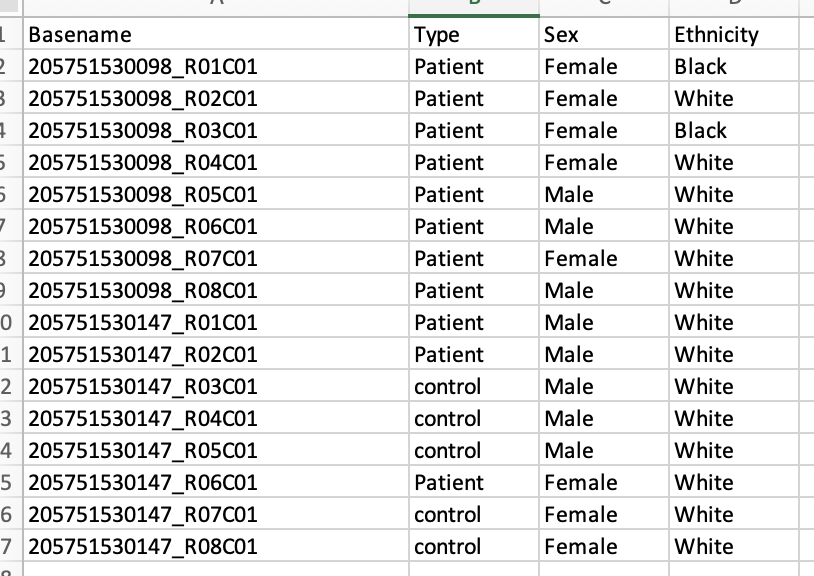
Hi I am new to using bioinformatics to analyse methylation array data, I am trying to get my data set into a data frame to create a summarized experiment. I have used the read.metharray.sheet function, however I keep getting an error that I am not sure how to resolve.
targets <- read.metharray.sheet(dataDirectory,pattern = run_1) Error in read.metharray.sheet(dataDirectory, pattern = run_1) : 'base' does not exists
This is the csv file and the idat files that I am using.
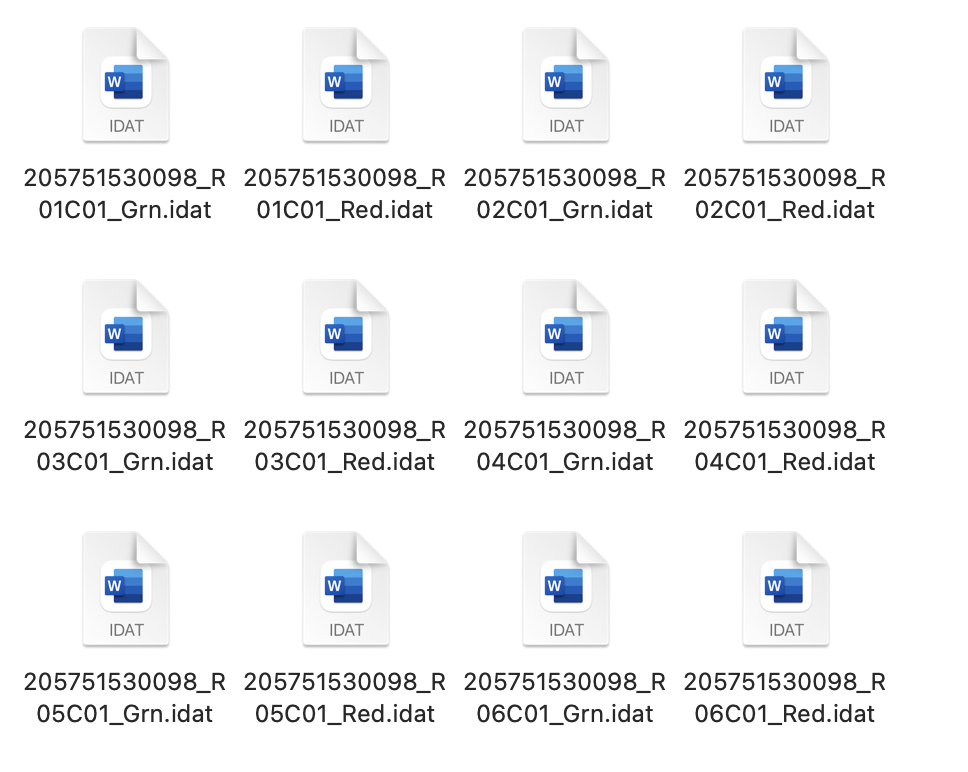
Any help is appreciated thank you!,