Hello,
I have an issue with my RNAseq regarding the variance between my biological replicates.
If i include them as a factor of variance in my design, i can fix the issue nicely and looking at the PCA plot when removing"batcheffect" with:
mat <- assay(vsd)
mat <- limma::removeBatchEffect(mat, vsd$Biological_rep)
assay(vsd) <- mat
the samples cluster very nicely according to their different treatment conditions.
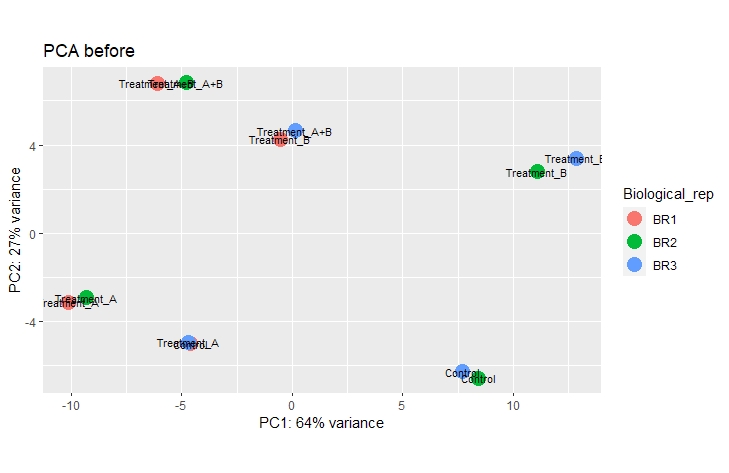
My question now is if it is acceptable to do this or if this takes away the whole point of using replicates in the first place.
Thanks alot for your help and insight!
Lukas

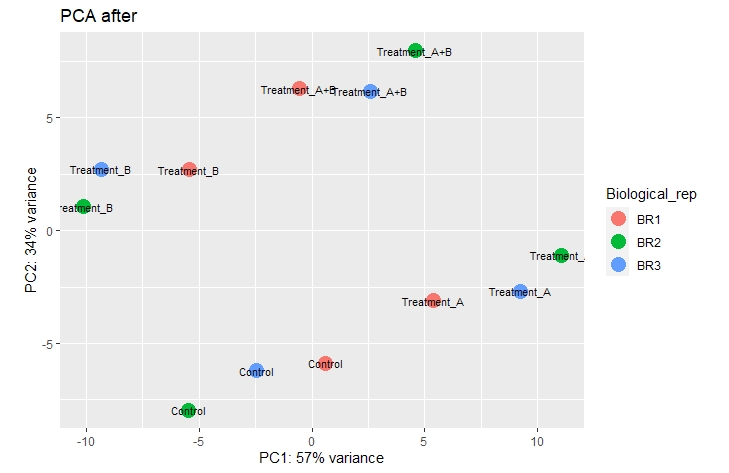

Thank you very much for your quick reply.
By including it into my model you would mean adding it to the design like this?