Even for a multi-exon knockout you could (and indeed we often do in our KO mice) still detect some borderline expression. I mean, look at your data, you have a strong reduction of reads, almost two powers of 10, so that is fine. After all it's the functional protein knockout that counts, anyway the RNA-seq counts will be highly significant.
Yes, it is evident that the gene's expression is reduced. But I just want to figure out why there are still some counts? It is due to the knock out technology or RNA-seq technology?
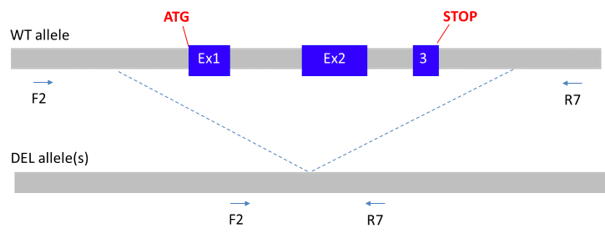
How was the gene knocked out? If they just introduced a stop codon into it, transcription could still happen.
Even for a multi-exon knockout you could (and indeed we often do in our KO mice) still detect some borderline expression. I mean, look at your data, you have a strong reduction of reads, almost two powers of 10, so that is fine. After all it's the functional protein knockout that counts, anyway the RNA-seq counts will be highly significant.
Yes, it is evident that the gene's expression is reduced. But I just want to figure out why there are still some counts? It is due to the knock out technology or RNA-seq technology?
The gene was totally knockedout by CRISPR/cas9.