Hi,
I am trying to plot a volcano plot using custom colors and I'd like to use legend labels containing subscript e.g. log2.
When I use colCustom argument, legendLabels argument does not work and use of expression function in named vector is not supported.
- Do you know if there is a way to replace the names of keyvals by legend labels in order to be able to use the expression function and add subscripts to legend text?
- Is it possible to hide/remove the legend completely from the plot?
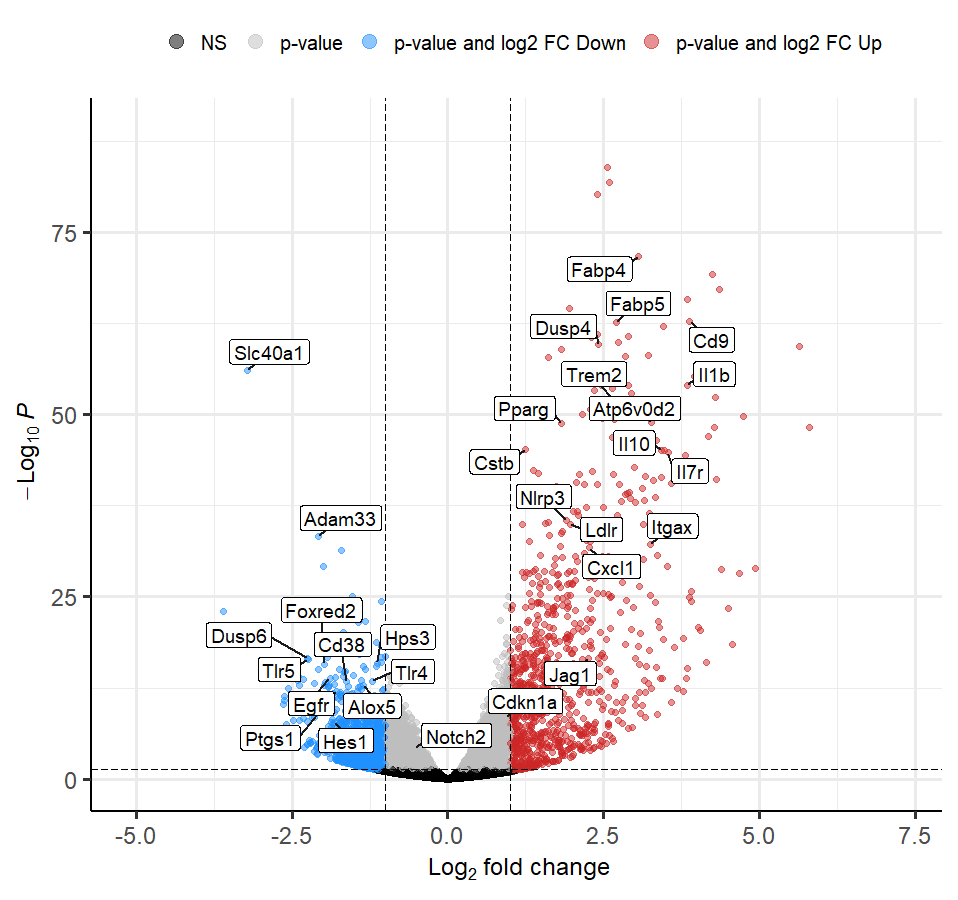
I used the following code to create custom colors and plot:
# create custom key-value pairs for 'high', 'low', 'mid' expression by fold-change
# this can be achieved with nested ifelse statements
keyvals <- ifelse(res$pvalue <= 0.05,
ifelse(res$log2FoldChange < -1,
'dodgerblue',
ifelse(res$log2FoldChange > 1,
'firebrick3',
'grey')),
ifelse(res$log2FoldChange < -1,
'black',
ifelse(res$log2FoldChange > 1,
'black',
'black'))
)
keyvals[is.na(keyvals)] <- 'grey'
names(keyvals)[keyvals == 'dodgerblue'] <- "p-value and log2 FC Down"
names(keyvals)[keyvals == 'grey'] <- 'p-value'
names(keyvals)[keyvals == 'firebrick3'] <- "p-value and log2 FC Up"
names(keyvals)[keyvals == 'black'] <- "NS" #'low & non sign'
names(keyvals)[keyvals == 'black'] <- "NS" #'mid & non sign'
names(keyvals)[keyvals == 'black'] <- "NS" #'high & non sign'
x = EnhancedVolcano(res,
lab = res$Genes,
x = 'log2FoldChange',
y = 'pvalue',
selectLab = genes_to_draw,
FCcutoff = 1,
pCutoff = 0.05,
colCustom = keyvals,
# col = c("yellow", "green", "grey", "firebrick3"), # non-sign/nonFC, non-sign/FC, sign/nonFC, sign/FC
drawConnectors = TRUE,
arrowheads = FALSE,
widthConnectors = 1,
lengthConnectors = unit(1, "cm"),
boxedLabels = TRUE,
# legend = c("NS", "p-value", "p-value and log2 FC Down", "p-value and log2 FC Up"),
# legendLabels = c('NS', 'p-value', expression(p-value~and~Log[2]~FC), expression(p-value~and~Log[2]~FC)),
legendLabSize = 15,
title = NULL, subtitle = NULL, caption = NULL)
print(x)
sessionInfo( )
R version 4.1.2 (2021-11-01)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=Greek_Greece.1253 LC_CTYPE=Greek_Greece.1253 LC_MONETARY=Greek_Greece.1253 LC_NUMERIC=C LC_TIME=Greek_Greece.1253
system code page: 1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] EnhancedVolcano_1.12.0 ggrepel_0.9.1 ggplot2_3.3.6
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-3 rjson_0.2.21 deldir_1.0-6 ellipsis_0.3.2 circlize_0.4.15 htmlTable_2.4.1
[8] GlobalOptions_0.1.2 base64enc_0.1-3 fs_1.5.2 clue_0.3-61 rstudioapi_0.14 farver_2.1.1 remotes_2.4.2
[15] fansi_1.0.3 codetools_0.2-18 splines_4.1.2 extrafont_0.18 doParallel_1.0.17 cachem_1.0.6 knitr_1.40
[22] pkgload_1.3.0 Formula_1.2-4 Rttf2pt1_1.3.11 cluster_2.1.2 png_0.1-7 shiny_1.7.2 compiler_4.1.2
[29] backports_1.4.1 assertthat_0.2.1 Matrix_1.5-1 fastmap_1.1.0 cli_3.4.1 later_1.3.0 htmltools_0.5.3
[36] prettyunits_1.1.1 tools_4.1.2 gtable_0.3.1 glue_1.6.2 dplyr_1.0.10 maps_3.4.0 Rcpp_1.0.9
[43] cellranger_1.1.0 vctrs_0.4.2 ggalt_0.4.0 extrafontdb_1.0 iterators_1.0.14 xfun_0.33 stringr_1.4.1
[50] ps_1.7.1 openxlsx_4.2.5 mime_0.12 miniUI_0.1.1.1 lifecycle_1.0.3 devtools_2.4.5 MASS_7.3-54
[57] scales_1.2.1 promises_1.2.0.1 parallel_4.1.2 proj4_1.0-11 RColorBrewer_1.1-3 ComplexHeatmap_2.13.1 memoise_2.0.1
[64] gridExtra_2.3 ggrastr_1.0.1 rpart_4.1-15 latticeExtra_0.6-30 stringi_1.7.8 S4Vectors_0.32.4 corrplot_0.92
[71] foreach_1.5.2 checkmate_2.1.0 BiocGenerics_0.40.0 pkgbuild_1.3.1 zip_2.2.1 shape_1.4.6 rlang_1.0.6
[78] pkgconfig_2.0.3 matrixStats_0.62.0 lattice_0.20-45 purrr_0.3.5 labeling_0.4.2 htmlwidgets_1.5.4 processx_3.7.0
[85] tidyselect_1.2.0 magrittr_2.0.3 R6_2.5.1 IRanges_2.28.0 generics_0.1.3 Hmisc_4.7-1 profvis_0.3.7
[92] DBI_1.1.3 pillar_1.8.1 foreign_0.8-81 withr_2.5.0 survival_3.2-13 nnet_7.3-16 ash_1.0-15
[99] tibble_3.1.8 crayon_1.5.2 interp_1.1-3 KernSmooth_2.23-20 utf8_1.2.2 urlchecker_1.0.1 jpeg_0.1-9
[106] GetoptLong_1.0.5 usethis_2.1.6 grid_4.1.2 readxl_1.4.1 data.table_1.14.4 callr_3.7.2 digest_0.6.29
[113] xtable_1.8-4 httpuv_1.6.6 stats4_4.1.2 munsell_0.5.0 beeswarm_0.4.0 vipor_0.4.5 sessioninfo_1.2.2


Hi,
First of all thank you for the great package and the quick response.
The trick with
legendPosition = "none"works fine, you may want to add it in the manual along with the ’top’, ’bottom’, ’left’, ’right’.As for the
parseLabels, I set it toTRUEbut when running theEnhancedVolcanofunction got the following error:The modified code used is as follows:
If I remove the
legendargument (the one that caused the error), it works but showing thenames(keyvals)aslegendLabels.Same behavior with the first option
Any ideas?