This error occurs when I use the read.metharray.exp function in the minfi package to read the methylation data. How can I solve it? Thanks!
baseDir1 <- "./GDCdata/TCGA-LUAD/harmonized/DNA_Methylation/Masked_Intensities"
targets1 <- read.metharray.sheet(baseDir1,
pattern = "sample_sheet.csv")
RGset1 <- read.metharray.exp(targets=targets1,
force = T)
Error: BiocParallel errors 1 remote errors, element index: 1 506 unevaluated and other errors first remote error: Error in readChar(con, nchars = n): (converted from warning) truncating string with embedded nuls Timing stopped at: 0.14 0.003 0.145
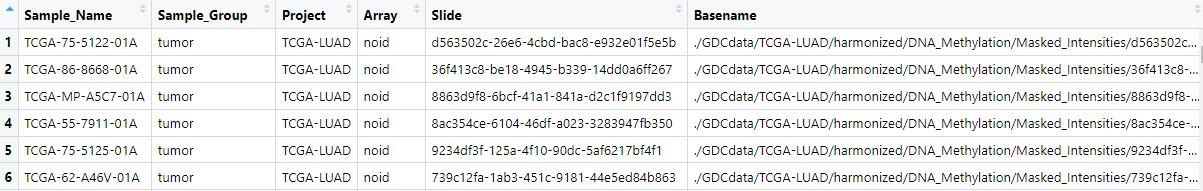
This is my targets1,
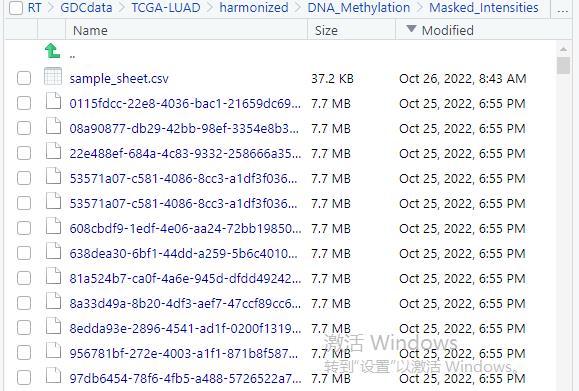
This is my idat file


I have similar issues, but it was not resolved after i followed the comments. Please kindly assist. Here is my line of code: I loaded target and manifest: targets <- read_excel("pheno_mouse_2023.xlsx") load("manifest.RData") temp <- data.frame(manifest)
betas <- openSesame(idat_dir, "MM285", manifest = temp)
Here is the error message
Error: BiocParallel errors 1 remote errors, element index: 1 59 unevaluated and other errors first remote error: Error in prepSesame(readIDATpair(x, platform = platform, manifest = manifest), : all(codes %in% cfuns$code) is not TRUE