Hi
I had a question about computationally singular matrices in DESeq and surrogate variables. After including all the SVs into my formula where I am analyzing paired samples (before/after treatment within a sample), I use the DESeq function and am returned with the error that my matrix is computationally singular. When I reduce the number of surrogate variables to 3, based on the n.sv() function using the "be" method, I no longer receive this error and am able to run the analysis. I want to know why the reduction of the number of SVs included in my design formula allows the dataset to run?
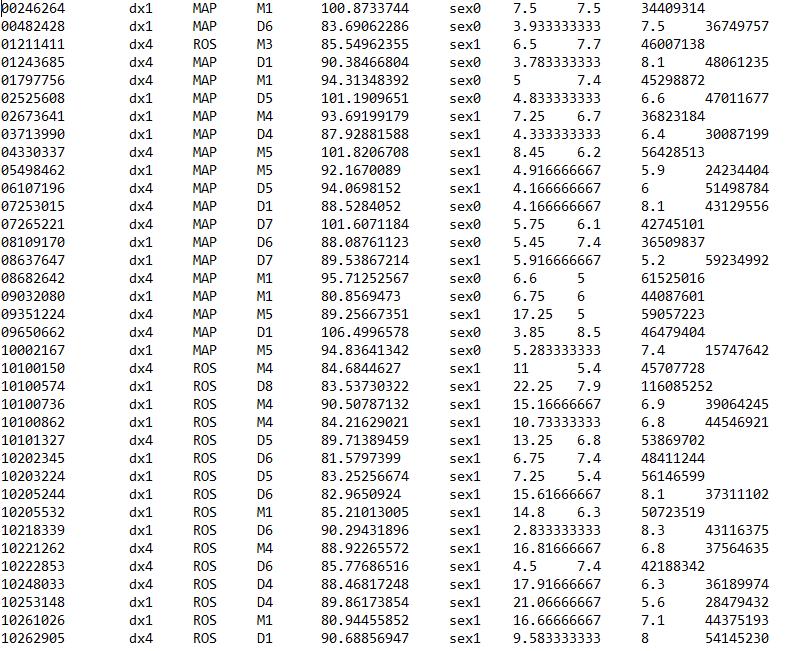
I am trying to create a design matrix that can be run in DESeq2 and have 8 variables that I am controlling for. The design matrix is as below:
dds <- DESeqDataSetFromMatrix(countData = countdata, colData = phenotype, design = ~ V3 + V4 + V5 + V6 + V7 + V8 + V9 + V2)
My design matrix is this:
Column headers are V1, V2.....V9. First column is ID, second column is treatment and the rest are what I am controlling for in respect to treatment. However, when I create the design matrix and run DESeq2, i get that error that my matrix is computationally singular. I have looked at similar questions posted here and checked if my design matrix has variables correlating highly with each other and that is not the case. I wanted to ask, first, that how many variables can I control for in DESEq2? Second, looking at my design matrix, what could be causing this error, Appreciate any help. Code should be placed in three backticks as shown below