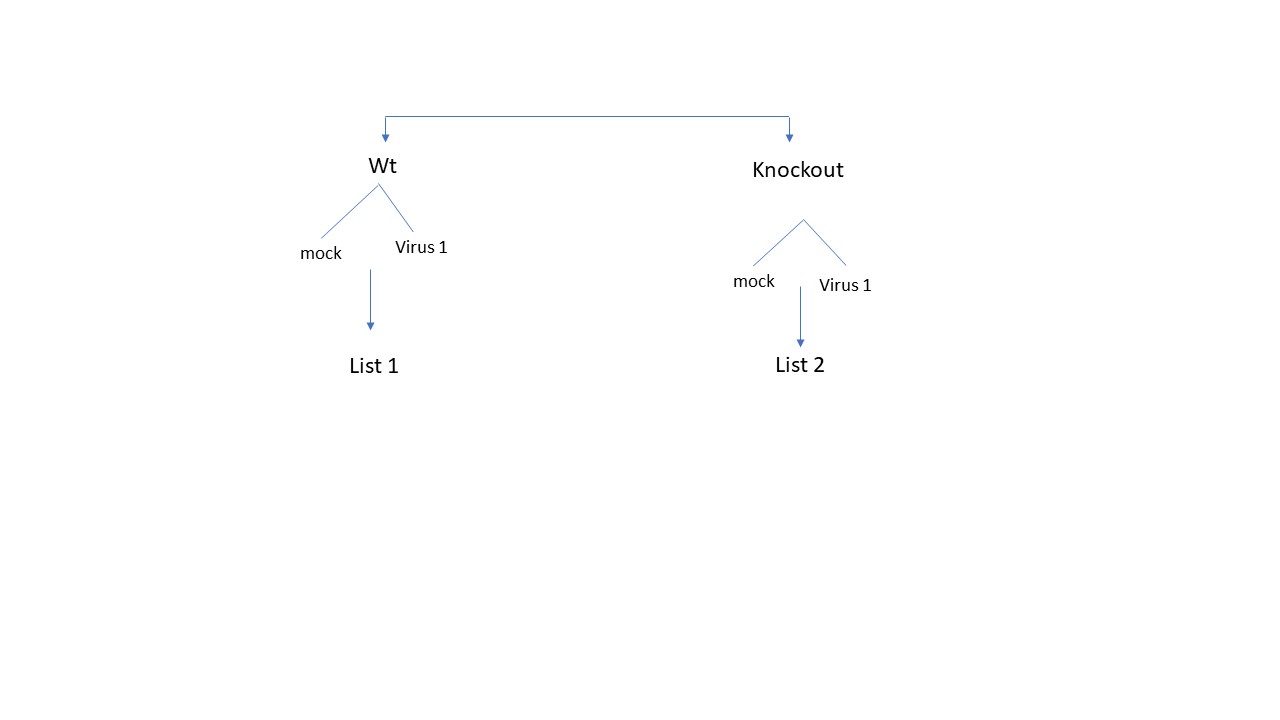
Dear All, I have two cell types (wild type and knockout), which is once is mock-treated and once is infected with a virus. I would like to know what are the differential genes that are a results of knockout after infection. The design looks like this
To answer this question, I have seen two scenario in many publication, but still confused :
To get the list of DE genes comparing Wt mock vs Wt infected with virus (list 1) [supposed to be the genes different due to infection of wild type)], and get also list 2 (comparing mock knockout vs virus infected knockout) [list of genes differ due to infection in knockout]., and then get unique genes that appeared only in list 2 is to directly compare virus infected wt cells vs virus infected knockout cells. Here I am skeptic if this is even valid, because the DE genes here is not solely due to knockout effect but also different cell background. Which one of these 2- scenario would reveal the most accurate gene list ?
Thanks


Cross-posted: https://www.biostars.org/p/9538149/ -- Folks suggested to use interactions, as described e.g. in the DESeq2/edgeR/limma manuals to quite some detail.