There's probably a more sophisticated way to do this, but here's a semi-brute force way, using KEGG's REST API.
> gnmap <- read.table("https://rest.kegg.jp/link/hsa/pathway/", sep = "\t")
> names(gnmap) <- c("Path","GeneID")
## Woo brute force
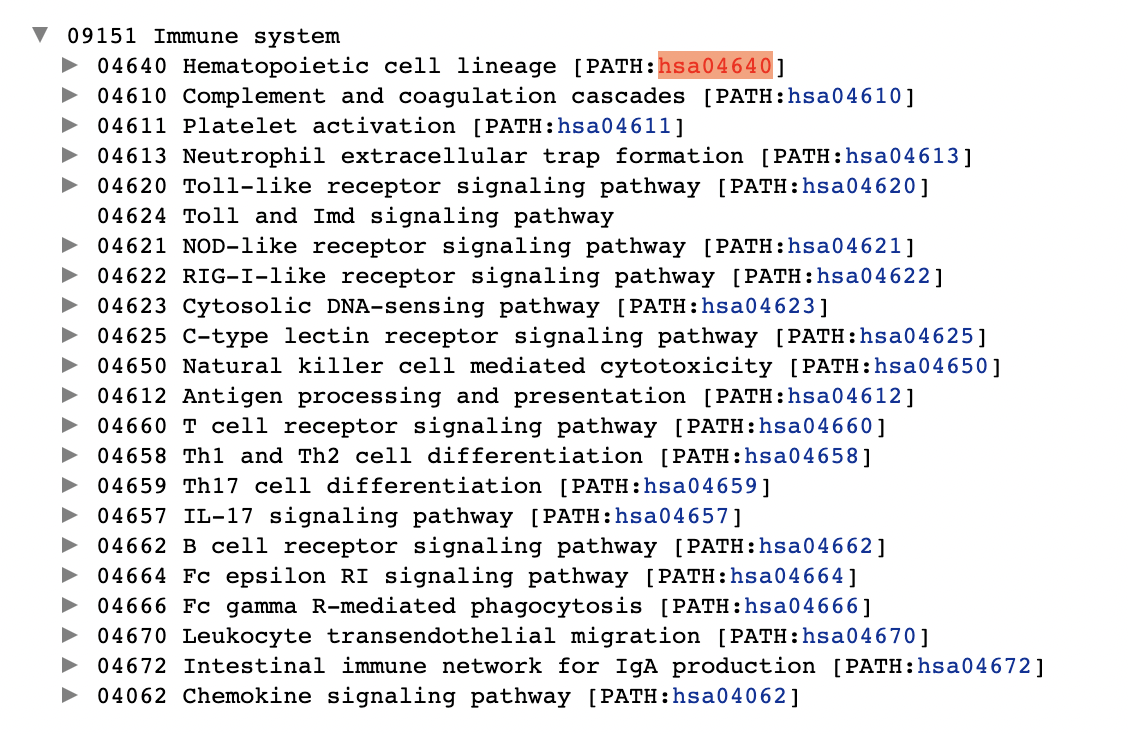
> whatIwant <- c(paste0("path:hsa046", c(40,10,11,13,20,24,21,22,23,25,60,12,60,58,59,57,62,64,66,70,72)), "path:hsa04062")
> gnmap2 <- subset(gnmap, Path %in% whatIwant)
> library(org.Hs.eg.db)
> gnmap2$Symbol <- mapIds(org.Hs.eg.db, gsub("hsa:", "", gnmap2$GeneID), "SYMBOL", "ENTREZID")
'select()' returned 1:1 mapping between keys and columns
> head(gnmap2)
Path GeneID Symbol
8506 path:hsa04062 hsa:10000 AKT3
8507 path:hsa04062 hsa:10235 RASGRP2
8508 path:hsa04062 hsa:10344 CCL26
8509 path:hsa04062 hsa:10451 VAV3
8510 path:hsa04062 hsa:10563 CXCL13
8511 path:hsa04062 hsa:10663 CXCR6
## Or maybe clean that up a bit.
> gnmap2$Path <- gsub("path:", "", gnmap2$Path)
> gnmap2$GeneID <- gsub("hsa:", "", gnmap2$GeneID)
> head(gnmap2)
Path GeneID Symbol
8506 hsa04062 10000 AKT3
8507 hsa04062 10235 RASGRP2
8508 hsa04062 10344 CCL26
8509 hsa04062 10451 VAV3
8510 hsa04062 10563 CXCL13
8511 hsa04062 10663 CXCR6


James' answer is great. Just for completeness, the
limmapackage has two helper functions that might be helpful: