Hi,
I am doing a meta analysis and ran into this peculiarity with SVA:
In at least one study (GSE147851), the first SV found by sva correlates entirely with the sample groups provided in mod1.
Could one of you kind people help me explain what is going on?
I ran SVA as follows. (The dds object is available here)
Metadata (for this example, I only used sample groups 1 and 3 of the original data)
> SummarizedExperiment::colData(dds)
DataFrame with 6 rows and 5 columns
study id source final_description group_nr
<character> <character> <character> <character> <factor>
GSM4447088 GSE147851 GSM4447088 ATRT patient tumor BT12,SMARCB1 1
GSM4447089 GSE147851 GSM4447089 ATRT patient tumor BT12,SMARCB1 1
GSM4447090 GSE147851 GSM4447090 ATRT patient tumor BT12,SMARCB1 1
GSM4447094 GSE147851 GSM4447094 ATRT patient tumor CHLA-06,SMARCB1 3
GSM4447095 GSE147851 GSM4447095 ATRT patient tumor CHLA-06,SMARCB1 3
GSM4447096 GSE147851 GSM4447096 ATRT patient tumor CHLA-06,SMARCB1 3
I normalized the counts of the samples of interest with DESeq2::normTransform. (varianceStabilizingTransformation had similar effect).
normalized_counts <- DESeq2::normTransform(dds) %>%
SummarizedExperiment::assay()
> colSums(normalized_counts) / 1e6
GSM4447088 GSM4447089 GSM4447090 GSM4447094 GSM4447095 GSM4447096
0.5111944 0.5137143 0.5227076 0.5441454 0.5557738 0.5531739
Defined mod0 and mod1 as
mod1 <- stats::model.matrix(~group_nr, data = metadata)
(Intercept) group_nr3
GSM4447088 1 0
GSM4447089 1 0
GSM4447090 1 0
GSM4447094 1 1
GSM4447095 1 1
GSM4447096 1 1
attr(,"assign")
[1] 0 1
attr(,"contrasts")
attr(,"contrasts")$group_nr
[1] "contr.treatment"
mod0 <- stats::model.matrix(~1, data = metadata)
(Intercept)
GSM4447088 1
GSM4447089 1
GSM4447090 1
GSM4447094 1
GSM4447095 1
GSM4447096 1
attr(,"assign")
[1] 0
And ran SVA
> svs <- sva::sva(normalized_counts, mod = mod1, mod0 = mod0)
Number of significant surrogate variables is: 2
Iteration (out of 5 ):1 2 3 4 5
> svs$sv
[,1] [,2]
[1,] 0.4726919 0.76011710
[2,] 0.2982086 -0.32400285
[3,] 0.4410821 -0.50232115
[4,] -0.3597558 -0.18110270
[5,] -0.4101281 0.09592035
[6,] -0.4420987 0.15138925
> cor(svs$sv[,1], mod1[,2])
[1] -0.9895797
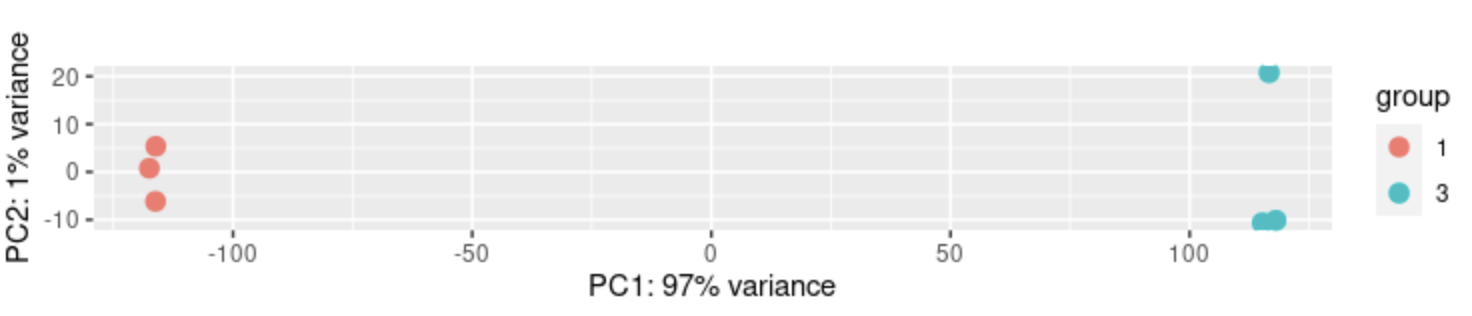
The samples are distributed as follows in PC space
DESeq2::plotPCA(DESeq2::normTransform(dds), intgroup = c("group_nr"))
> sessionInfo( )
R version 4.0.5 (2021-03-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
other attached packages:
[1] sva_3.44.0 BiocParallel_1.24.1
[3] genefilter_1.72.1 mgcv_1.8-38
[5] nlme_3.1-153 magrittr_2.0.1
[7] DESeq2_1.30.1 SummarizedExperiment_1.20.0
[9] Biobase_2.50.0 MatrixGenerics_1.2.1
[11] matrixStats_0.61.0 GenomicRanges_1.42.0
[13] GenomeInfoDb_1.26.7 IRanges_2.24.1
[15] S4Vectors_0.28.1 BiocGenerics_0.36.1
Edit: Tried fixing formatting