Hi,
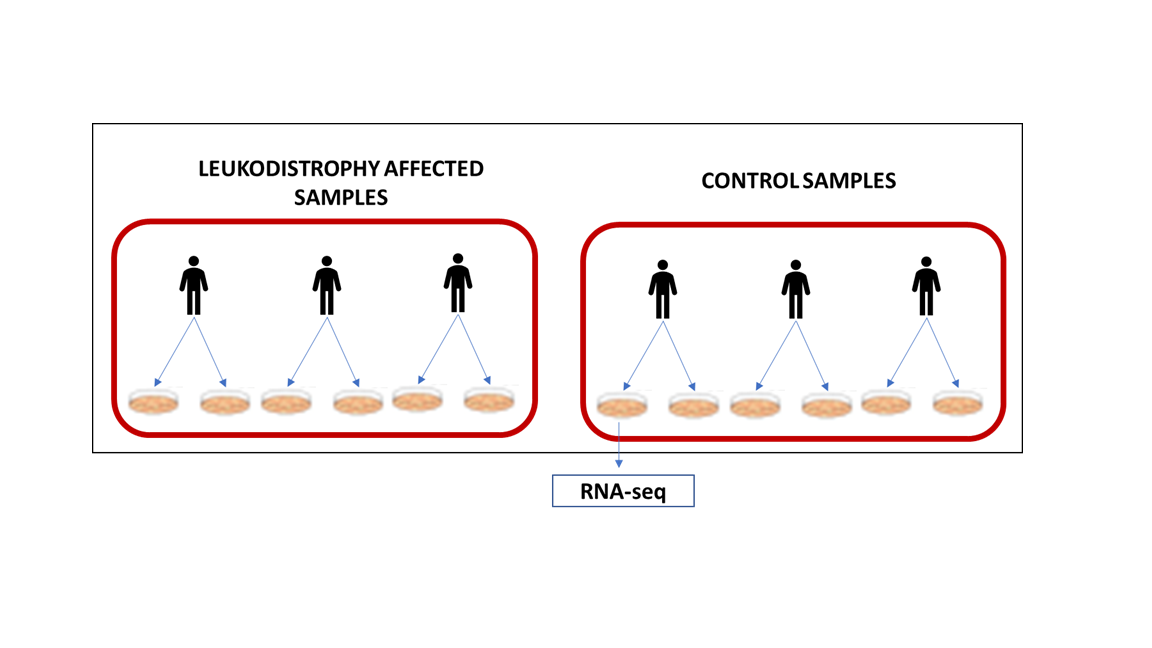
I am performing a DE analysis using Deseq2. For my experimental settings, I would like to compare leukodystrophy affected patient against control patient. For each condition, I have three patients and two replicates from the same patient, with a total of 6 replicates for each condition (see Figure attached).
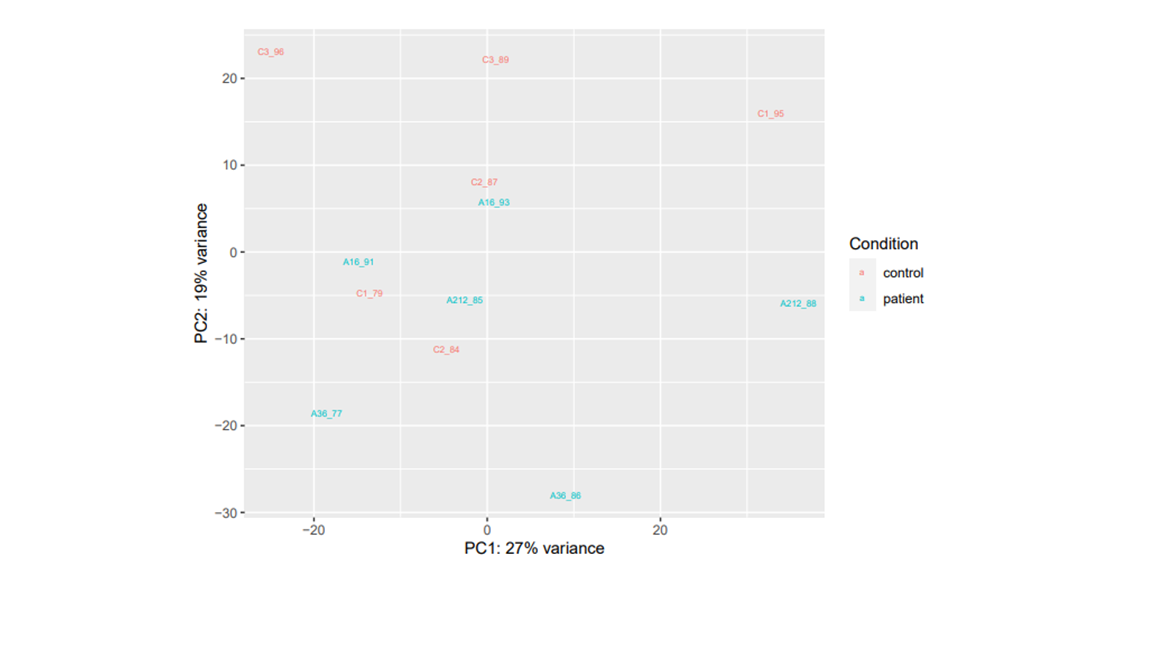
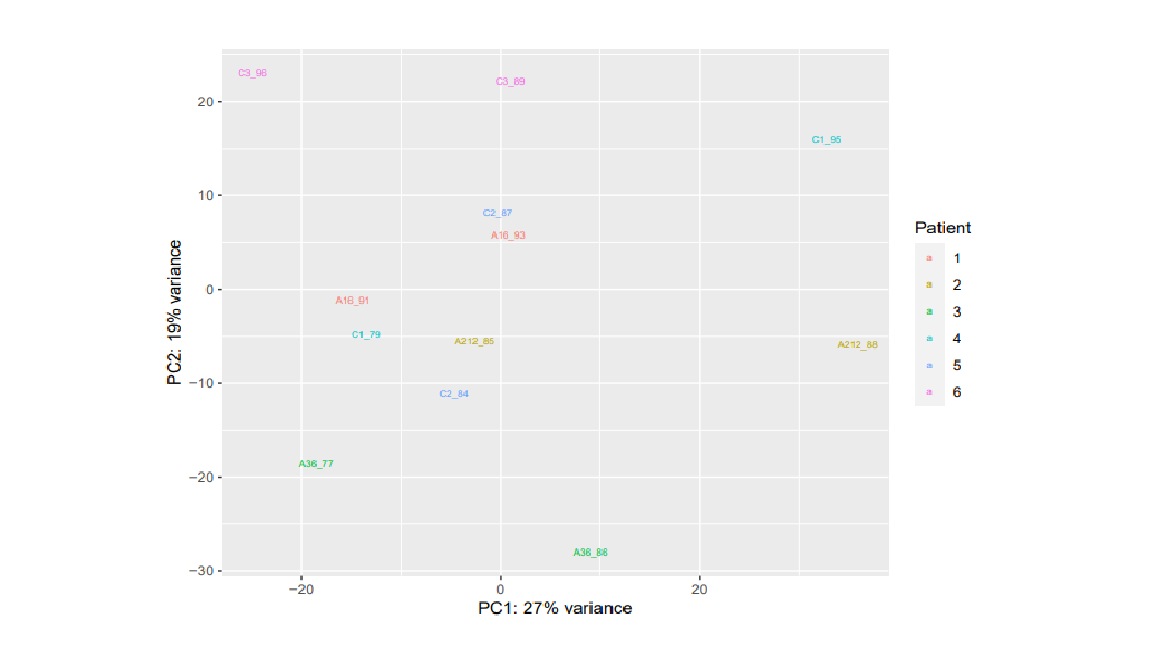
I am attaching also the sample distribution with PCA plots with condition variable and the information of patient
I was wondering if for this experiment it is recommended to perform a nested analysis or a “collapsed- replicates” analysis.
Best regards,
Giulia Lopatriello


Dear Michael,
I have run the analysis with limma and I have obtained no significant gene whereas in DESeq2 I have found approximately 100 DE genes.
Given these results, would you strongly recommend to use limma analysis with this type of experiment or could we use DESeq2?
Best regards,
Giulia L.
A key aspect of correct analysis is identifying the unit of analysis. If you want to compare across patients, you can't add replicates per patient without letting the model know that these are not i.i.d. observations. So yes, you will obtain FP by ignoring the correlations of these replicates, and you need to use duplicateCorrelation here.