I am trying to take the "difference of differences" in contrasts from two factors (sex and group). We have male and female animals (sex factor) that were untrained or trained for 1, 2, 4, or 8 weeks (group factor, i.e., "control", "1w", "2w", "4w", "8w"). I want to know the difference in the training effect at each time point between the two sexes.
Conceptually, this is what I want at the 1-week time point, for example:
(male:1w - male:control) - (female:1w - female:control)
However, I can't figure out how to do this in DESeq2. Specifying this contrast in limma would be straightforward, but I want to use DESeq2 for consistency with other arms of the differential analysis. I have looked at other answers here and here as well as the Interactions section of the DESeq2 tutorial, but I am still perplexed.
I have tried specifying the contrast with an interaction term (i.e. ~ sex*group) as well as with a new factor that pastes together the sex and group factors. I feel like I got the closest with the interaction term:
dds <- makeExampleDESeqDataSet(n=50,m=100)
dds$sex <- factor(rep(c("male","female"), each=25))
dds$group <- factor(rep(c("control","1w","2w","4w","8w"), 10))
# define reference levels
dds$sex <- relevel(dds$sex, ref="male")
dds$group <- relevel(dds$group, ref="control")
design(dds) <- ~ group*sex
dds <- DESeq(dds)
resultsNames(dds)
Which gives:
[1] "Intercept" "group_1w_vs_control" "group_2w_vs_control" "group_4w_vs_control" "group_8w_vs_control" "sex_female_vs_male" "group1w.sexfemale" "group2w.sexfemale"
[9] "group4w.sexfemale" "group8w.sexfemale"
From this post, I thought group1w.sexfemale would test if the 1-week effect (i.e., 1w - control) is different between females and males. However, the results aren't consistent with that. See below the plot of a gene that had a log fold-change of -23.2 and an adjusted p-value of 1e-4 for this specific contrast name. From the TMM-normalized data, I would expect a marginally significant result of roughly 3-fold.
I would appreciate any help!

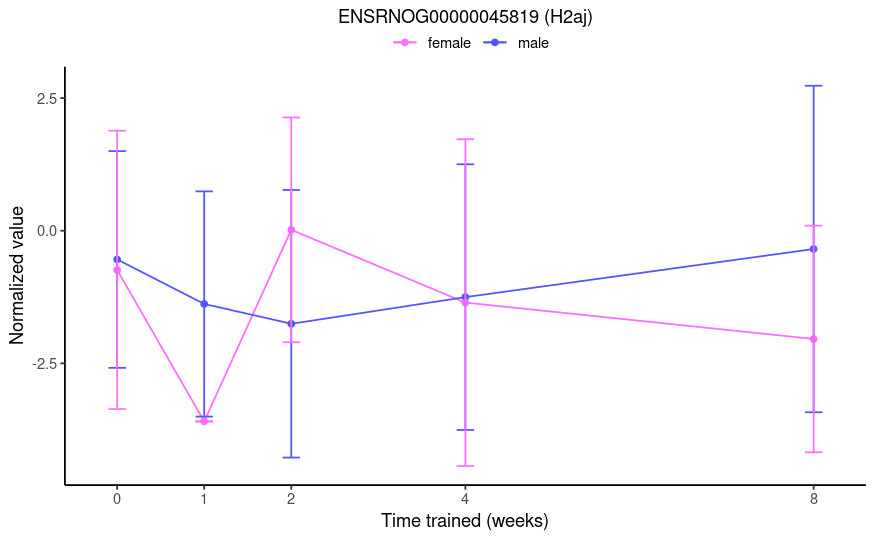

No, you misunderstood my question. This is not a simple comparison of two groups. I do not want to compare 1w F to 1w M. Rather, I want to compare the training effects at 1 week between the sexes, i.e.
(male:1w - male:control) - (female:1w - female:control).Right, that's exactly the comparison I want. But the results are not interpretable as such. If you look at the 0-week (control) and 1-week time points in the plot I provided, you will see that a logFC of -23.2 is not a reasonable result. Therefore, I do not believe that the
group1w.sexfemalecontrast is giving me the difference of differences that I want. Though perhaps the values for this gene are 0-inflated, giving me a much different logFC than I would expect with the TMM-normalized data.If you know how to formulate the result in limma, use a numeric-style
contrastwhen callingresults().E.g. if you want (D - C) - (B - A), then use a cell means design (no intercept) and then you will have coefficients A,B,C,D. Then you can specify
contrast=c(1, -1, -1, 1).Thank you, Michael Love - that did it! The results make sense now. From my original post, could you clarify what the
group1w.sexfemalecontrast would test? From reading the docs, I thoughtgroup1w.sexfemalewould be equivalent to(group_female_1w - group_female_control) - (group_male_1w - group_male_control), but they give very different results.They give a correlation of 1 for me, maybe look over the code.
Thank you! I'll look into it on my end.