Hi all,
I am uncertain about constructing a design matrix for my study below,
Our comparison would be contr.matrix <- makeContrasts(Condition = Infertile-Healthy,levels = colnames(design)).
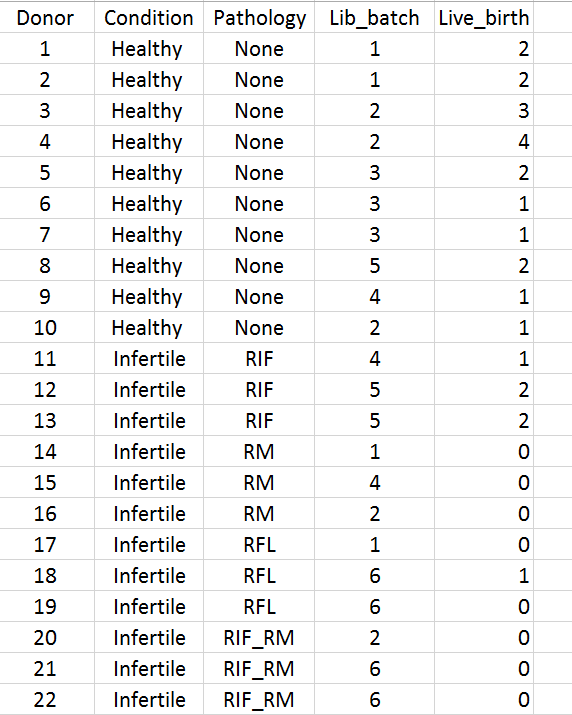
We are interested in the average differences driven by human fertility, hopefully taking into consideration of the infertile subpopulations (4 levels: RIF, RM, RFL and RIF_RM), and because PCA shows Lib_batch accounts for the highest proportion of variance in the data (followed by Pathology) so that will be accounted for.
However, all the healthy donors have no associated pathologies so design <- model.matrix(~0 + Condition + Lib_prep + Pathology, data = dgeList$samples) would give columns that are linearly dependent/not of full rank. We saw no significant differential expression when pathology is not accounted for, and we understand the sample size for each subpopulation is too small to be assessed independently. Would you be able to offer some advice in regard to constructing an appropriate design matrix for this study?
Thanks very much!!
Cheers, A


Thanks Gordon, that actually helps! I included
Pathologyin the design matrix and formed a contrast to find the difference between the average of 4 pathology groups and healthy and we saw some interesting findings!